Difference between revisions of "GBrowse/tool data"
m (slight updates) |
m |
||
| (8 intermediate revisions by the same user not shown) | |||
| Line 8: | Line 8: | ||
| full_name = Generic Genome Browser | | full_name = Generic Genome Browser | ||
| status = mature | | status = mature | ||
| − | | dev = | + | | dev = active |
| support = active | | support = active | ||
| − | | type = | + | | type = Genome Visualization & Editing |
| − | | | + | | platform = web |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| logo = GBrowseLogo.png | | logo = GBrowseLogo.png | ||
| home = http://gbrowse.org | | home = http://gbrowse.org | ||
| Line 35: | Line 31: | ||
Note that the information on this page refers to GBrowse 2; GBrowse 1.x is recommended only for applications where legacy browser support is required and a single database is used. | Note that the information on this page refers to GBrowse 2; GBrowse 1.x is recommended only for applications where legacy browser support is required and a single database is used. | ||
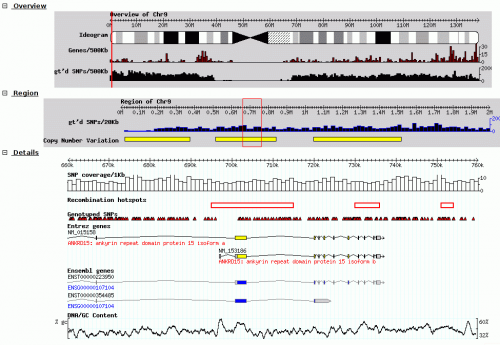
| − | | screenshot = [[image:GBrowse_screenshot1.png|none|thumb| | + | | screenshot = [[image:GBrowse_screenshot1.png|none|thumb|500px|GBrowse running on [http://hapmap.ncbi.nlm.nih.gov/downloads/index.html HapMap.org] [[Media:GBrowse_screenshot1.png|Click to view at full resolution]]]] |
| − | | public_server = | + | | public_server = |
| dl = | | dl = | ||
| dl_url = http://sourceforge.net/projects/gmod/files/Generic%20Genome%20Browser/ | | dl_url = http://sourceforge.net/projects/gmod/files/Generic%20Genome%20Browser/ | ||
| Line 42: | Line 38: | ||
| dl_src_url = | | dl_src_url = | ||
| dl_dev = | | dl_dev = | ||
| + | GBrowse can also be found on [http://search.cpan.org CPAN] and in the Debian sid ("unstable") apt package repository and can be installed via apt-get in Ubuntu 12.04 and later. | ||
| dl_dev_url = https://github.com/GMOD/GBrowse | | dl_dev_url = https://github.com/GMOD/GBrowse | ||
| getting_started_preamble = | | getting_started_preamble = | ||
| Line 96: | Line 93: | ||
* {{GBrowse_pod|ORACLE_AND_POSTGRESQL}} | * {{GBrowse_pod|ORACLE_AND_POSTGRESQL}} | ||
| − | | papers = * Using the Generic Genome Browser ([[GBrowse]]). <ref name=PMID:19957275/> | + | | papers = |
| + | * Using the Generic Genome Browser ([[GBrowse]]). <ref name=PMID:19957275/> | ||
* SNP@Evolution: a hierarchical database of positive selection on the human genome. <ref name=PMID:19732458/> ([[GBrowse]]-related) | * SNP@Evolution: a hierarchical database of positive selection on the human genome. <ref name=PMID:19732458/> ([[GBrowse]]-related) | ||
* TBrowse: an integrative genomics map of Mycobacterium tuberculosis. <ref name=PMID:19683474/> ([[GBrowse]]-related) | * TBrowse: an integrative genomics map of Mycobacterium tuberculosis. <ref name=PMID:19683474/> ([[GBrowse]]-related) | ||
| Line 106: | Line 104: | ||
| presentations = | | presentations = | ||
| tutorials = | | tutorials = | ||
| − | + | ; [[GBrowse Tutorial|GBrowse tutorial from 2012 GMOD Summer School]]. This relies heavily on the {{GBrowseAdminTutorialLink| GBrowse Administration Tutorial}}. | |
| − | + | : Demonstrates setting up, configuring and using [[GBrowse]] with some sample data. GBrowse is provided on an Amazon Machine Image; see [[Cloud|GMOD in the Cloud]] for more information on getting a GMOD AMI. | |
| − | + | ; [[GBrowse Tutorial 2010|GBrowse tutorial from 2010 GMOD Summer School]] | |
| − | + | : Set up and run [[GBrowse]] with sample data. It provides a VMware image to work on, and relies heavily on the {{GBrowseAdminTutorialLink|GBrowse Administration Tutorial}}. | |
| + | ; {{GBrowseAdminTutorialLink|GBrowse Administration Tutorial}} | ||
| + | : Step by step guide on how to configure and load data into [[GBrowse]]. Administration tutorials are available for both the {{GBrowseAdminTutorialLink|current version}}, and the earlier {{GBrowse1AdminTutorialLink|1.x versions}}. | ||
| + | ;[http://search.cpan.org/dist/GBrowse/htdocs/general_help.html Usage tutorial] | ||
| + | :GBrowse usage tutorial | ||
| + | ; [[GBrowse NGS Tutorial]] | ||
| + | : Instructions on how to visualize [[next generation sequencing]] data in GBrowse using [http://samtools.sourceforge.net SAMtools]. The tutorial includes a starting VMware image, and uses the example data that comes with SAMtools. | ||
| + | ; [http://youtu.be/jxA6VMN97Y8 GBrowse video tutorial] | ||
| + | : Produced by [http://eupathdb.org EuPathDB]; please direct praise and thanks to them! | ||
| + | ; [http://www.openhelix.com/gbrowse GBrowse User Tutorial] at [http://www.openhelix.com OpenHelix] | ||
| + | : A Flash based tutorial on using GBrowse. Provided by [http://www.openhelix.com OpenHelix]. Tutorial includes slides, handouts and exercises. NB: this tutorial is for [[GBrowse 1.x]] | ||
| wild_urls = | | wild_urls = | ||
*[http://www.wormbase.org/tools/genome/gbrowse/c_elegans/ WormBase] | *[http://www.wormbase.org/tools/genome/gbrowse/c_elegans/ WormBase] | ||
| Line 119: | Line 127: | ||
Please report bugs to the [http://sourceforge.net/tracker/?func=add&group_id=27707&atid=391291SourceForge Bug Tracker] (select 'Category: Gbrowse'). | Please report bugs to the [http://sourceforge.net/tracker/?func=add&group_id=27707&atid=391291SourceForge Bug Tracker] (select 'Category: Gbrowse'). | ||
| − | | logo_info = The [[: | + | | logo_info = The [[:File:GBrowseLogo.png|GBrowse logo]] was created by [mailto:alexisnb1@yahoo.com Alex Read], a participant in the [[Spring 2010 Logo Program]], while a design student at [http://www.linnbenton.edu Linn-Benton Community College]. |
| dev_ppl = | | dev_ppl = | ||
| − | | dev_status = | + | | dev_status = |
| − | | contact_email = | + | | contact_email = lincoln.stein@gmail.com; scott@scottcain.net |
| − | | input = | + | | input = [[GFF3]], [[GFF2]] |
| output = | | output = | ||
| see_also = [[JBrowse]], the successor to GBrowse, built with JavaScript for a faster, more interactive user experience; and [[WebGBrowse]], a tool for configuring GBrowse. | | see_also = [[JBrowse]], the successor to GBrowse, built with JavaScript for a faster, more interactive user experience; and [[WebGBrowse]], a tool for configuring GBrowse. | ||
| demo_server = | | demo_server = | ||
| survey_link = GBrowse | | survey_link = GBrowse | ||
| + | |release date=2001 | ||
| + | | extras = | ||
| + | {{GitcComponent}} | ||
| + | | | ||
}} | }} | ||
Latest revision as of 18:35, 10 September 2013
This page stores the data that populates the GBrowse wiki page.
{{ {{{template}}}
| name = GBrowse
| full_name = Generic Genome Browser
| status = mature
| dev = active
| support = active
| type = Genome Visualization & Editing
| platform = web
| logo = GBrowseLogo.png
| home = http://gbrowse.org
| about = GBrowse is a combination of database and interactive web pages for manipulating and displaying annotations on genomes. Features include:
- Simultaneous bird's eye and detailed views of the genome.
- Scroll, zoom, center.
- Use a variety of premade glyphs or create your own.
- Attach arbitrary URLs to any annotation.
- Order and appearance of tracks are customizable by administrator and end-user.
- Search by annotation ID, name, or comment.
- Supports third party annotation using GFF formats.
- Settings persist across sessions.
- DNA and GFF dumps.
- Connectivity to different databases, including BioSQL and Chado.
- Multi-language support.
- Third-party feature loading.
- Customizable plug-in architecture (e.g. run BLAST, dump & import many formats, find oligonucleotides, design primers, create restriction maps, edit features)
Note that the information on this page refers to GBrowse 2; GBrowse 1.x is recommended only for applications where legacy browser support is required and a single database is used.
| screenshot =| public_server = | dl = | dl_url = http://sourceforge.net/projects/gmod/files/Generic%20Genome%20Browser/ | dl_src = | dl_src_url = | dl_dev = GBrowse can also be found on CPAN and in the Debian sid ("unstable") apt package repository and can be installed via apt-get in Ubuntu 12.04 and later. | dl_dev_url = https://github.com/GMOD/GBrowse | getting_started_preamble = | req = GBrowse is Perl-based and the GBrowse 2.x modules are hosted on CPAN. GBrowse can be installed using the standard Perl module build procedure, or automated using a network-based install script. In order to use the net installer, you will need to have Perl 5.8.6 or higher and the Apache web server installed. | install =
| config =
| doc =
- GBrowse FAQ
- Annotation Help
- Balloons:
- GBrowse Persistent Variables
- GBrowse img
- Glyphs and Glyph Options
- Rubber Band Selection
- Gbrowse Benchmarking
- GBrowse User Uploads
POD documentation
There are many useful POD documents included with the distribution. These are converted to HTML files when you install the package, and can be found in /gbrowse/docs/pod.
GBrowse 2.x pod documents can also be viewed online at CPAN:
- FAQ
- INSTALL
- INSTALL.MacOSX
- README-chado
- README-gff-files (see also GFF)
- README-lucegene
- BIOSQL_ADAPTER_HOWTO
- GENBANK_HOWTO
- PLUGINS_HOWTO
- DAS_HOWTO
- MAKE_IMAGES_HOWTO
- GBROWSE_IMG
- ORACLE_AND_POSTGRESQL
| papers =
- Using the Generic Genome Browser (GBrowse). [1]
- SNP@Evolution: a hierarchical database of positive selection on the human genome. [2] (GBrowse-related)
- TBrowse: an integrative genomics map of Mycobacterium tuberculosis. [3] (GBrowse-related)
- FishMap: a community resource for zebrafish genomics. [4] (GBrowse-related)
- Using the Generic Genome Browser (GBrowse). [5]
- Genome-Wide Analysis of Nucleotide-Level Variation in Commonly Used Saccharomyces cerevisiae Strains. [6] (See YSB.)
- Gbrowse Moby: a Web-based browser for BioMoby Services. [7]
- The generic genome browser (GBrowse): a building block for a model organism system database. [8] PDF
| presentations = | tutorials =
- GBrowse tutorial from 2012 GMOD Summer School. This relies heavily on the GBrowse2 Admin Tutorial.
- Demonstrates setting up, configuring and using GBrowse with some sample data. GBrowse is provided on an Amazon Machine Image; see GMOD in the Cloud for more information on getting a GMOD AMI.
- GBrowse tutorial from 2010 GMOD Summer School
- Set up and run GBrowse with sample data. It provides a VMware image to work on, and relies heavily on the GBrowse2 Admin Tutorial.
- GBrowse2 Admin Tutorial
- Step by step guide on how to configure and load data into GBrowse. Administration tutorials are available for both the GBrowse2 Admin Tutorial, and the earlier 1.x versions.
- Usage tutorial
- GBrowse usage tutorial
- GBrowse NGS Tutorial
- Instructions on how to visualize next generation sequencing data in GBrowse using SAMtools. The tutorial includes a starting VMware image, and uses the example data that comes with SAMtools.
- GBrowse video tutorial
- Produced by EuPathDB; please direct praise and thanks to them!
- GBrowse User Tutorial at OpenHelix
- A Flash based tutorial on using GBrowse. Provided by OpenHelix. Tutorial includes slides, handouts and exercises. NB: this tutorial is for GBrowse 1.x
| wild_urls =
| mail =
| Mailing List Link | Description | Archive(s) | |
|---|---|---|---|
| GBrowse & GBrowse_syn | gmod-gbrowse | GBrowse and GBrowse_syn users and developers. | Gmane, Nabble (2010/05+), Sourceforge |
| gmod-gbrowse-cmts | Code updates. | Sourceforge |
Please report bugs to the Bug Tracker (select 'Category: Gbrowse').
| logo_info = The GBrowse logo was created by Alex Read, a participant in the Spring 2010 Logo Program, while a design student at Linn-Benton Community College. | dev_ppl = | dev_status = | contact_email = lincoln.stein@gmail.com; scott@scottcain.net | input = GFF3, GFF2 | output = | see_also = JBrowse, the successor to GBrowse, built with JavaScript for a faster, more interactive user experience; and WebGBrowse, a tool for configuring GBrowse. | demo_server = | survey_link = GBrowse |release date=2001 | extras =
Included in
|
}}
Cite error: <ref> tags exist, but no <references/> tag was found