GMOD
Artemis-Chado Integration Tutorial

This Artemis-Chado Integration tutorial was presented by Robin Houston, Tim Carver and Giles Velarde at the 2009 GMOD Summer School - Europe, August 2009. The most recent Artemis tutorial can be found at the Artemis Tutorial page.
This tutorial walks you through how to use the Artemis annotation editor with a Chado database.
VMware
.
To run ACT use the act script:
./act -Dchado="localhost:5432/chado_pathogen?gmod" -Dibatis
From the ‘File’ menu select the option ‘Open Database and SSH File Manager’ and login. Drag and drop the Plasmodium entries from the Database Manager into the ACT selection window. Also, drag and drop the comparison files into this window, so it looks something like this (note the featureId numbers may well be different as these are the Chado feature_id):
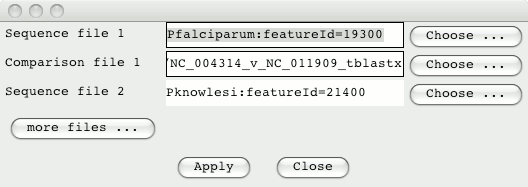
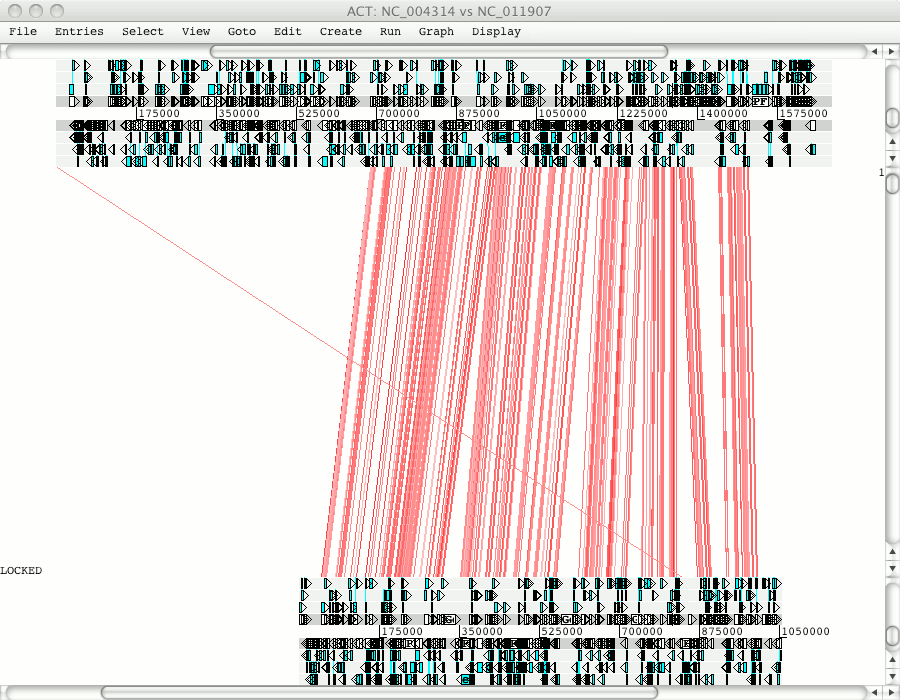
Click on Apply to read these entries and open up ACT. You can use the right hand scroll bar to zoom in and out. If you zoom out you can indentify the regions that match between these sequences.
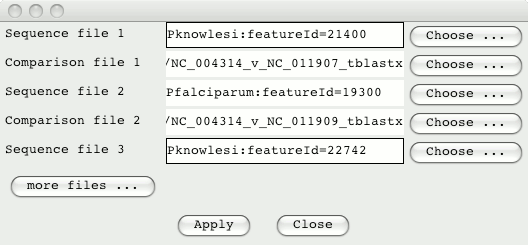
ACT can display multiple pairwise comparison. So the two P.knowlesi
sequences can be compared to the P.falciparum sequence. From the ACT
launch window go to the File menu and select ‘Open Database and SSH File
Manager’. Drag in the sequences and comparison files (clicking on ‘more
files’ to add the additional sequence and comparison).
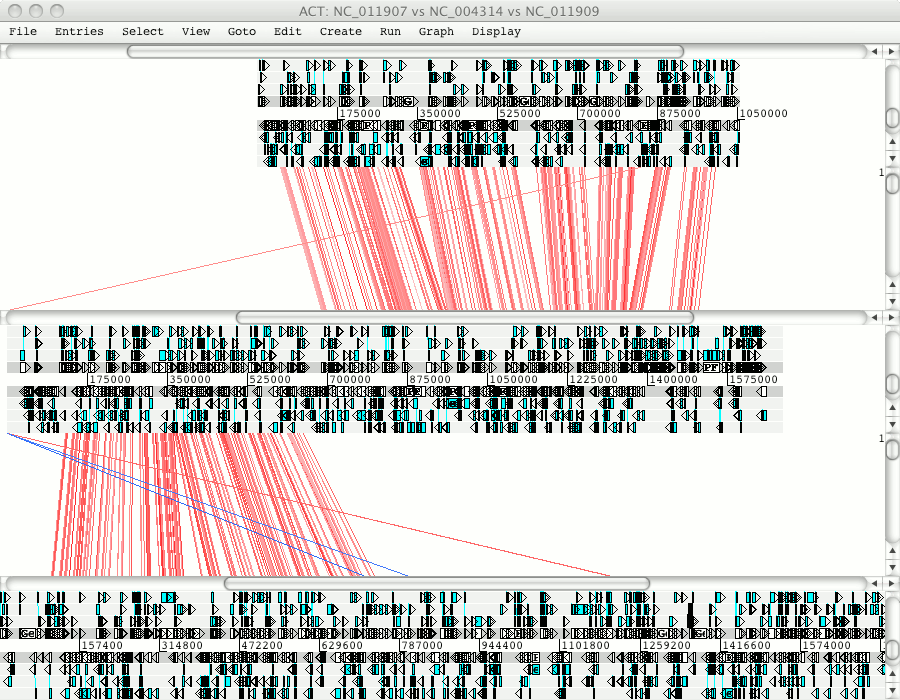
Zooming out you will see that Pfalciparum chromosome 10 matches to regions in Pknowlesi chromosome 7 and 9.
Writing Out Sequence Files
Artemis can write out EMBL and GFF files for an entry
opened from the database. You can optionally flatten the gene model
(i.e. gene, transcript, exon) to just a CDS feature. Also an option is
given to ignore any obsolete features. For EMBL it uses mappings for
conversion of the keys and qualifiers. These mappings are stored in the
etc/key_mapping and etc/qualifier_mapping files.
A utility script (etc/write_db_entry) is also provided as a means of
writing out multiple sequences from the database. The script takes the
following options:
-h show help
-f [y|n] flatten the gene model, default is y
-i [y|n] ignore obsolete features, default is y
-s space separated list of sequences to read and write out
-o [EMBL|GFF] output format, default is EMBL
-a [y|n] for EMBL submission format change to n, default is y
Try running:
etc/writedb_entry -Dchado="localhost:5432/chado_pathogen?gmod" NC_004314
Mailing List
There is an Artemis mailing list: artemis-user.
References
Facts about “Artemis-Chado Integration Tutorial”
| Has topic | Artemis + |