GMOD
Tripal Tutorial (v1.0)

This Tripal tutorial was presented by Stephen Ficklin at the 2012 GMOD Summer School, August 2012. The most recent Tripal tutorial can be found at the Tripal Tutorial page.
Welcome to the Tripal v1.0 Tutorial. Here you will find instructions for installation, usage and administration of a Tripal-based genome website.
Note: An updated tutorial for Tripal v1.1 can be found here
To follow along with the tutorial, you will need to use AMI ID: ami-b7fa4dde, name: GMOD 2012 day 2 start, available in the US East (N. Virginia) region. See the GMOD Cloud Tutorial for information on how to get this AMI.
Contents
- 1 What is Tripal
- 2 Content Management System
- 3 Drupal
- 4 Chado
- 5 Goals of Tripal
- 6 Structure of Tripal
- 7 Sites Running Tripal
- 8 Resources
- 9 Contributing Organizations
- 10 Funding
- 11 Pre-planning
- 12 Server Installation
- 13 Database Setup
- 14 Install Drupal
- 15 Explore Drupal
- 16 Prepare Drupal for Tripal
- 17 Drush
- 18 Tripal Installation
- 19 Using
Tripal
- 19.1 Creating Organism Pages
- 19.2 Creating an Analysis
- 19.3 Creating a Database Cross Reference
- 19.4 Loading Feature Data
- 19.5 Creating Feature Pages
- 19.6 Materialized Views
- 19.7 Feature Page Configuration
- 19.8 Loading Functional Data Using Extension Modules
- 19.9 Customizing The Look-and-Feel of Tripal
- 19.10 Adding Additional Resources
- 19.11 Advanced Features
What is Tripal
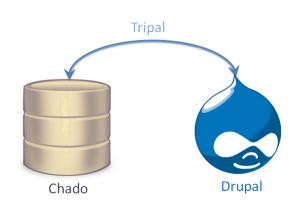
Tripal is a suite of PHP5 modules that bridges the Drupal Content Managment System (CMS) and GMOD Chado. The goal is to simplify construction of a community genomics website to enable individual labs or research communities to construct a high-quality, standards-based website for data sharing and collaboration.
Content Management System
Definition From Wikipedia:
A content management system (CMS) is the collection of procedures used to manage work flow in a collaborative environment. These procedures can be manual or computer-based. The procedures are designed to do the following:
- Allow for a large number of people to contribute to and share stored data
- Control access to data, based on user roles (defining which information users or user groups can view, edit, publish, etc.)
- Aid in easy storage and retrieval of data
- Reduce repetitive duplicate input
- Improve the ease of report writing
- Improve communication between users
In a CMS, data can be defined as nearly anything: documents, movies, pictures, phone numbers, scientific data, and so forth. CMSs are frequently used for storing, controlling, revising, semantically enriching, and publishing documentation. Serving as a central repository, the CMS increases the version level of new updates to an already existing file. Version control is one of the primary advantages of a CMS.
Drupal
Drupal is an open-source freely available CMS with thousands of users and existing sites. Features of Drupal
- A well-supporting community.
- Books, tutorials and online forums for help .
- Hundreds of user-contributed extension modules that are freely available.
- Hundreds of user-contributed themes to instantly change the look-and-feel of the site
- User management infrastructure.
- Allows for non-coding manipulation of the website contents. Anyone can edit content.
- Easy to install and maintain
Drupal website: http://www.drupal.org Drupal modules: http://www.drupal.org/project/modules Drupal themes: http://www.drupal.org/project/themes
Tripal v1.0 is compatible with Drupal v6. This is the final Drupal 6 compatible version. Later releases will be compatible with Drupal v7.
Chado
You can find more detailed information about Chado here: http://gmod.org/wiki/Chado_-_Getting_Started. However, one thing to remember in regards to Tripal organization is that Chado has a modular structure:
- Audit - for database audits
- Companalysis - for data from computational analysis
- Contact - for people, groups, and organizations
- Controlled Vocabulary (cv) - for controlled vocabularies and ontologies
- Expression - for summaries of RNA and protein expresssion
- General - for identifiers
- Genetic - for genetic data and genotypes
- Library - for descriptions of molecular libraries
- Mage - for microarray data
- Map - for maps without sequence
- Organism - for taxonomic data
- Phenotype - for phenotypic data
- Phylogeny - for organisms and phylogenetic trees
- Publication (pub) - for publications and references
- Sequence - for sequences and sequence features
- Stock - for specimens and biological collections
- WWW -
Tripal is also modular along these same designations.
Goals of Tripal
- Simplify Construction of Biological Databases
- Reduce time of development
- Reduce costs
- Reduce technical resources (i.e. programmers, systems admins).
- A non-technical site administrator can add content without knowing PHP, HTML, JavaScript.
- Greater Flexibility of the Biological Website
- Social Networking
- Non-biological content
- Outreach, tutorials, documentation, protocols, publications
- Expandability
- Site can be programmatically expanded in any way
- Changes to base-code are not needed but modules are added.
- Availability of an Application Programmer Interface (API)
- Reusability
- All code can be shared. Expansion modules created by one group can be shared with all.
Structure of Tripal
Tripal is a collection of modules that integrate with Drupal. These modules are divided into hierarchical categories:
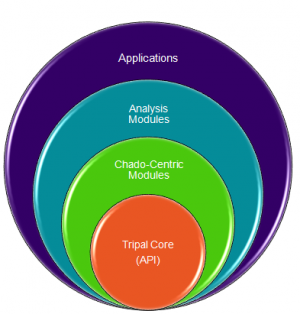
The Tripal Core level contains supportive functionality for all other modules. It contains
- A jobs management utility
- A utility to manage materialized views
- An API for these features
- Functions for managing module specific CV terms
- Functions for interfacing with Chado.
The Chado-centric modules provide:
- Edit/Update/Delete for Chado modules.
- Bulk loaders for these data
- Basic visualizations for data in Chado specific for the module
- An API for easily accessing Chado.
Analysis modules provide
- Custom visualization for specific analyses (e.g. Blast, KEGG, InterProScan, Unigene construction)
- Uses the API from the Tripal Analysis (Chado-centric) module.
Applications:
- These are full blown applications that use Tripal, Drupal and Chado and typically consist of several Chado-centric modules, Analysis modules and custom built modules. (e.g. Breeders Toolbox currently under construction).
Sites Running Tripal
| Site Name | URL | Tripal Version |
|---|---|---|
| Pulse Crops Genomics & Breeding | <a href=”http://knowpulse2.usask.ca/portal/” class=”external free” | |
| rel=”nofollow”>http://knowpulse2.usask.ca/portal/</a> | development version | |
| Genome Database for Vaccinium | <a href=”http://www.vaccinium.org” class=”external free” | |
| rel=”nofollow”>http://www.vaccinium.org</a> | v0.3.1b | |
| Genome Database for Rosacaee | <a href=”http://www.rosaceae.org” class=”external free” | |
| rel=”nofollow”>http://www.rosaceae.org</a> | v1.0 (under construction) | |
| Cool Season Food Legume Database | <a href=”http://www.gabcsfl.org” class=”external free” | |
| rel=”nofollow”>http://www.gabcsfl.org</a> | v0.3.1b | |
| Cacao Genome Database | <a href=”http://www.cacaogenomedb.org” class=”external free” | |
| rel=”nofollow”>http://www.cacaogenomedb.org</a> | pre v1.0 | |
| Fagaceae Genome Web | <a href=”http://www.fagaceae.org” class=”external free” | |
| rel=”nofollow”>http://www.fagaceae.org</a> | v0.2 | |
| Citrus Genome Database | <a href=”http://www.citrusgenomedb.org” class=”external free” | |
| rel=”nofollow”>http://www.citrusgenomedb.org</a> | v0.2 | |
| Marine Genomics Project | <a href=”http://www.marinegenomics.org” class=”external free” | |
| rel=”nofollow”>http://www.marinegenomics.org</a> | pre v0.1 |
Resources
The Tripal Sourceforge home site where you can find everything about Tripal: http://tripal.sourceforge.net
GMOD Tripal mailing lists: http://gmod.org/wiki/GMOD_Mailing_Lists
GMOD Tutorials from previous GMOD schools: http://gmod.org/wiki/Tripal
Contributing Organizations
Individuals from these organizations have provided design and coding for Tripal v1.0
![]()
![]()
Acknowledgments are extended to the Clemson University Genomics Institute where Tripal was first conceived and where development of earlier releases was performed.
![]()
Also, special thanks are extended to the GMOD project for logistical support and community interaction!!
Funding
Funding for Tripal v1.0 has been provided through various grants from various sources. For a complete list, please see the recent publication:
Stephen P. Ficklin, Lacey-Anne Sanderson, Chun-Huai Cheng, Margaret Staton, Taein Lee, Il-Hyung Cho, Sook Jung, Kirstin E Bett, Dorrie Main. Tripal: a construction Toolkit for Online Genome Databases. Database, Sept 2011. Vol 2011.
Pre-planning
IT Infrastructure
Tripal requires a server with adequate resources to handle the expected load and systems administration skills to get the machine up and running, applications installed and everything properly secure. Tripal requires a PostgreSQL databases server, Apache (or equivalent) web server, PHP5 and several configuration options to make it all work. However, once these prerequisites are met, working with Drupal and Tripal are quite easy.
There are three basic ways you could get a Tripal/Drupal/Chado database web server available for your site
- Option #1 In-house dedicated servers: You may have access to servers in your own department or group which you have administrative control and wish to install Tripal/Drupal/Chado on these.
- Option #2 Institutional IT support: Your institution may provide IT servers and would support your efforts to install a website with database backend.
- Option #3 Commercial web-hosting: If options #1 and #2 are not available to you, commercial web-hosting is an affordable option. For large databases you may require a dedicated server.
After selection of one of the options that works best for you you can arrange your database/webserver in the following ways:
- Arrangement #1: The database and web server are housed on a single server. This is the approach taken by this course. It is necessary to gain access to a machine with enough memory (RAM), hard disk speed and space, and processor power to handle both services.
- Arrangement #2: The database and web server are housed on different servers. This provides dedicated resources to each service (i.e. web and database).
Selection of an appropriate machine
Databases are typically bottlenecked by RAM and disk speed. Selection of the correct balance of RAM, disk speed, disk size and CPU speed is important and dependent on the size of the data. Unfortunately, we have only tested production installations of Tripal on a limited number of server configurations. The best advice is to consult an IT professional who can recommend a server installation tailored for the expected size of your data.
Note: Tripal does require command-line access to the web server with adequate local file storage for loading of large data files. Be sure to check with your service provider to make sure command-line access is possible.
Technical Skills
Depending on your needs, you may need additional Technical support….
Tripal already supports my data, what personnel to I need to maintain it?
- Someone to install/setup the IT infrastructure
- Someone who understands the data to load it properly
Tripal does not yet support all of my data, but I want to use what’s been done and expand on it….?
- Someone to install/setup the IT infrastructure
- Someone who understands the data to load it properly
- PHP/HTML/CSS/JavaScript programmer(s) to write your custom extensions
Why Use Tripal
Tripal v1.0 does not yet fully support Chado in terms of visualization of all primary data types. It does support all of Chado in terms of data access. So why use Tripal?
- If you want to use Chado
- You need a web interface
- You need CMS capabilities (distributed content editing, user management, social networking… i.e. Drupal)
- You want to contribute to a community effort to help build a tool others can use.
- Participate in a community of other database developers using the same technology, confronting similar problems and helping each other.
- It is all open-source and free technology!
Server Installation
The following instructions are for setup of Tripal on an Ubuntu version 12.04 server edition. When possible, alternative command-line statements have been added to this tutorial as users of other Linux version have provided feedback. Unless specifically identified, all commands are for Ubuntu 12.04 linux.
During installation of the Ubuntu 12.04 server please select the following software packages for installation:
- OpenSSH server
- LAMP (includes Apache and PHP)
- PostgreSQL database
After installation the Ubuntu Unity Desktop can be installed. For the virtual machine image that accompanies this tutorial, the following command was issued to install the desktop:
sudo apt-get install ubuntu-desktop
Reboot your server after installation of the Ubuntu Desktop.
Apache Setup
Apache is the web server software. Apache should be installed. On the Ubuntu server, navigate to your new website using this address: http://localhost/. You should see the text “It works!”.
Enable the rewrite module for apache. This is useful so that we can use Clean URLs with Drupal. Clean URLs are not required but make the page URLs easier to use. We’ll discuss clean URLs later.
cd /etc/apache2/mods-enabled
sudo ln -s ../mods-available/rewrite.load
Next we need to edit the apache configuration file to give Drupal full controls of options within the directory root. Edit the /etc/apache2/sites-available/000-default file:
cd /etc/apache2/sites-available/
sudo gedit default
And change the AllowOverride option from None to All:
<Directory /var/www/>
Options Indexes FollowSymLinks MultiViews
AllowOverride All
Order allow,deny
allow from all
</Directory>
Now restart your apache again.
sudo /etc/init.d/apache2 restart
Setup PHP
PHP comes loaded onto the server, but we need a few other packages:
 First install php5 using
Ubuntu’s apt-get utility:
First install php5 using
Ubuntu’s apt-get utility:
sudo apt-get install php5-pgsql
sudo apt-get install php5-gd
![]() For Suse Linux you may need
to install the php-posix package:
For Suse Linux you may need
to install the php-posix package:
yum install php-posix
 For RedHat
Linux you may also need to install the php-process package:
For RedHat
Linux you may also need to install the php-process package:
yum install php-process
Change some php settings (as root):
cd /etc/php5/apache2
sudo gedit php.ini
Set the memory_limit to something larger than 128M (should not
exceed physical memory, be conservative but not too much so):
memory_limit = 2048M;
Now, restart the webserver:
sudo /etc/init.d/apache2 restart
Install phpPgAdmin
phpPgAdmin is a nice web-based utility for easy administration of a PostgreSQL database. It is not required for successful operation of Tripal but is very useful.
sudo apt-get install phppgadmin
Now navigate to the URL [http://localhost/phppgadmin] and you should see the following:
Before continuing, we want to password protect PhpPgAdmin using Apache’s access control mechanisms. To do this, we will create two hidden files in the /usr/share/phppgadmin directory. First we need to instruct apache to use password protection for PhpPgAdmin. To do this, edit the phppgadmin config file for apache:
cd /etc/apache2/conf.d
sudo gedit phppgadmin
Within the Directory stanza add the following:
AuthType Basic
AuthName "Password Required"
AuthUserFile /usr/share/phppgadmin/.htpasswd
Require User tripaladmin
Note: it is necessary that the preceding lines appear below the ‘Directory /usr/share/phppgadmin’ line.
The lines above instruct apache to use basic authentication, that the password file is located at /usr/share/phppgadmin/.htpasswd and the only user allowed to login is ‘tripaladmin’. Next we need to create the password file:
cd /usr/share/phppgadmin
sudo htpasswd -c .htpasswd tripaladmin
The htpasswd command above will create the .htpasswd file and add the new user ‘tripaladmin’. You will need to set a password when requested.
Now, restart the webserver:
sudo /etc/init.d/apache2 restart
A password should now be required when accessing the PhpPgAdmin page.
Database Setup
Drupal can run on a MySQL or PostgreSQL database but Chado prefers PostgreSQL so that is what we will use for both Drupal and Chado. We need to create the Drupal database. The following command can be used to create a new database user and database.
First, become the ‘postgres’ user:
sudo su - postgres
Next, create the new ‘drupal’ user account. This account will not be a “superuser’ nor allowed to create new roles, but should be allowed to create a database.
createuser -P drupal
When requested, enter an appropriate password:
Enter password for new role: *********
Enter it again: *********
Shall the new role be a superuser? (y/n) n
Shall the new role be allowed to create databases? (y/n) y
Shall the new role be allowed to create more new roles? (y/n) n
Finally, create the new database:
createdb drupal -O drupal
We no longer need to be the postgres user so exit
exit
Install Drupal
Software Installation
We want to install Drupal into our web document root (/var/www). We
want to be able to do this as our ‘ubuntu’ user. So, first, let’s set
the directory permissions to allow this:
cd /var
sudo chgrp -R ubuntu www
sudo chmod -R g+rw www
In the command above we set the group of the directory to be ubuntu (our user group) and we gave the directory read and write permissions for the group.
Tripal currently requires version 6.x of Drupal. Drupal can be freely downloaded from the http://www.drupal.org website. At the writing of this Tutorial the most recent version of Drupal 6 is version 6.26. The software can be downloaded manually from the Drupal website through a web browser or we can use the ‘wget’ command to retrieve it:
cd /var/www
wget http://ftp.drupal.org/files/projects/drupal-6.26.tar.gz
Note: The current version of Drupal is Drupal 7.x. Relase v1.0 of Tripal is the final version that will be compatible with Drupal 6.x. Future major releases of Tripal will be compatible with Drupal 7.x.
Next, we want to install Drupal. We will use the tar command to uncompress the software:
cd /var/www
tar -zxvf drupal-6.26.tar.gz
Notice that we now have a drupal-6.26 directory with all of the Drupal
files. We want the Drupal files to be in our document root, not in a
‘drupal-6.26’ subdirectory. So, we’ll move the contents of the directory
up one level:
mv drupal-6.26/* ./
mv drupal-6.26/.htaccess ./
mv index.html index.html.orig
Note: It is extremely important the the hidden file .htaccess is
also moved (note the second ‘mv’ command above. Check to make sure this
file is there
ls -l .htaccess
Notice that the last of the three mv commands renames the index.html
file and calls it index.html.orig. The index.html file was serving
as the home page for the website. Drupal uses an index.php page for
it’s home page but the web server has preference for the index.html
page. So, we move it out of the way.
Configuration File
Next, we need to tell Drupal how to connect to our database. To do this we have to setup a configuration file. Drupal comes with an example configuration file which we can borrow.
First navigate to the location where the configuration file should go:
cd /var/www/sites/default/
Next, copy the example configuration that already exists in the
directory to be our actual configuration file by renaming it to
settings.php.
cp default.settings.php settings.php
Now, we need to edit the configuration file to tell Drupal how to connect to our database server. To do this we’ll use an easy to use text editor gedit
gedit settings.php
Find the variable $db_url and set it to this
$db_url = 'pgsql://drupal:********@localhost/drupal';
Replace the text ‘********’ with your database password for the user ‘tripal’ created previously.
Final directory creation
Finally, we need to create three new directories. The first is the
files directory which Drupal uses for storing uploaded files.
cd /var/www/sites/default
mkdir files
sudo chown ubuntu:www-data files
sudo chmod g+rw files
The above command creates the directory but sets the group to be the web server (i.e. www) with read/write permissions. This way the web server can write to the directory but so can we.
Also, we need to create two new directories, one for storing module files we’ll be installing and another for themes which we’ll also be installing later:
Now create the modules and themes directory
cd /var/www/sites/all
mkdir modules
mkdir themes
Compatibility with other tools
We want to ensure that our Drupal installation doesn’t interfere with
other web-based tools, such as GBrowse. We need update a setting in the
.htaccess file that came with Drupal which instructs the web server to
look for both index.php and index.html files when serving pages.
Use ‘gedit’ to modify the /var/www.htaccess file.
cd /var/www
gedit .htaccess
Locate the line DirectoryIndex and change it to mach the following:
DirectoryIndex index.php index.html
Web-based Steps
Navigate to the installation page of our new web site http://localhost/install.php
Click the link in the middle section that reads Install Drupal in English
When the progress bar shows completing the page will switch to a configuration page with some final settings.
Set the following
- Site Name: Tripal Demo
- Site email: Your email address
- Administrator Username: administrator (all lower-case)
- Administrator Email: Your email address
- Password: ********
- Default time zone: leave as is
- Clean URLs: enabled
- Update Notification: check for updates automatically
Now, click the Save and Continue button. You will see a message about unable to send an email. This is safe to ignore as email capabilities are not fully enabled on this VMWare image. Now, your site is enabled. Click the link Your new site:
Drupal Cron Entry
The last step for installing Drupal is setting up the automatted Cron entry. The Drupal cron is used to automatically execute necessary housekeeping tasks on a regular interval. Cron is a UNIX facility for scheduling jobs to run at specific intervals.
Drupal itself requires an entry in the crontab to function. To edit the cron launch the crontab editor:
sudo crontab -e
A word on text editors such as nano.
Add this line to the crontab
0,30 * * * * /usr/bin/wget -O - -q http://localhost/cron.php > /dev/null
Now save the changes. We have now added a UNIX cron job that will occur
every 30 minutes that will execute the cron.php script and cause
Drupal to perform housekeeping tasks.
Explore Drupal
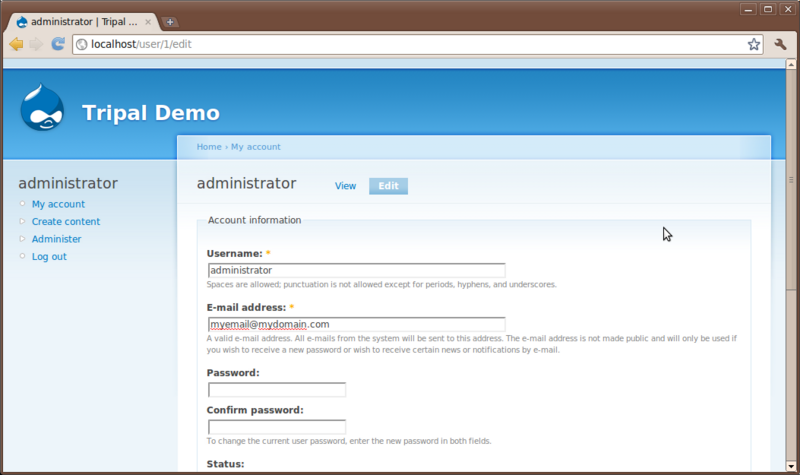
User Account Page
All users have an account page. Currently, we are logged in as the administrator. The account page is simple for now. Click the My account link on the left sidebar. You’ll see a brief history for the user and an Edit tab. Users can edit their own information using the edit interface:
Creating Content
Creation of content in Drupal is very easy. Click the Create content link on the left sidebar.
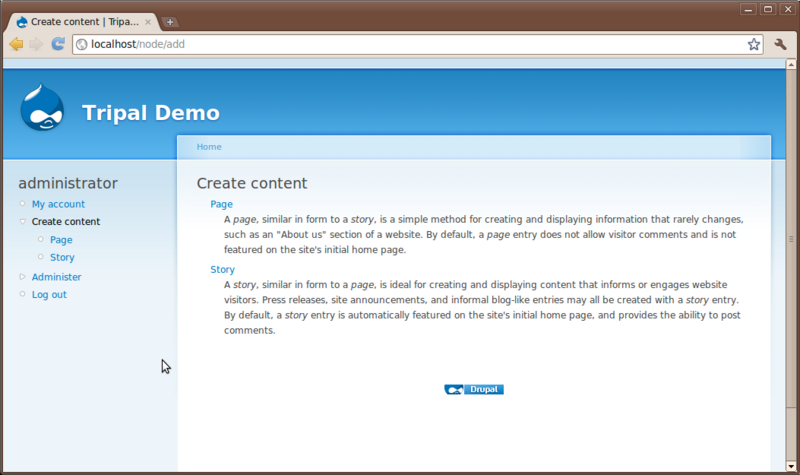
You’ll see two content types that come default with Drupal: Page and Story. Here is where a user can add simple new pages to the website without knowledge of HTML or CSS. Click the Page content type to see the interface for creating a new page:
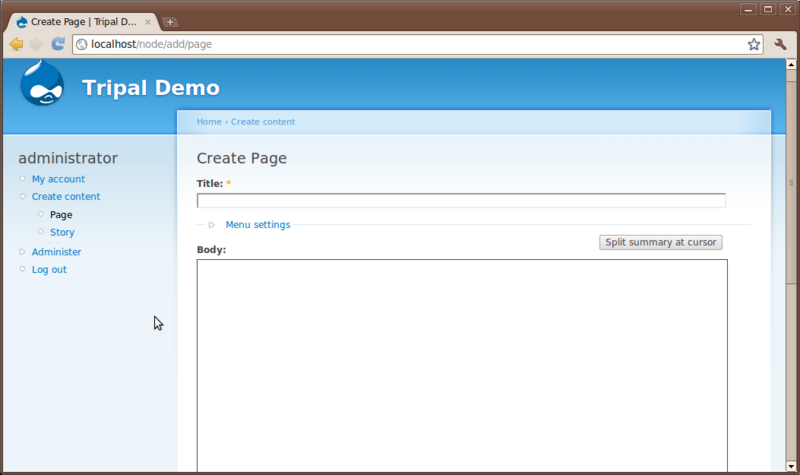
You’ll notice at the top a Title field and a Body text box. All pages require a title and typically have some sort of content entered in the body. Additionally, there are other options that allow someone to enter HTML if they would like, save revisions of a page to preserve a history and to set authoring and publishing information.
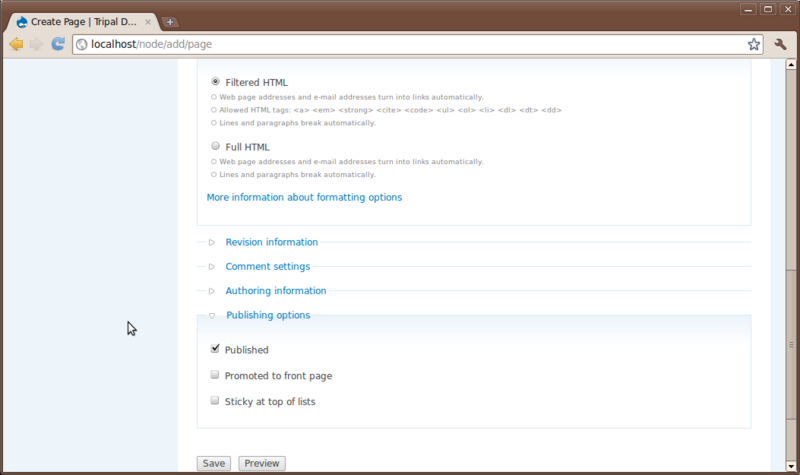
For practice, try to create two new pages. A Home page and an About page for our site. First create the home page and second create the about page. Add whatever text you like for the body.
Site Administration
Content Management
There are many options under the Administer link on the left sidebar. Here you can manage the site setup, monitor and control content, manage users and view reports.
We will not explore all of the options here but will visit a few of the more important ones for this tutorial. First, click the Content Management link on the left sidebar. You’ll see different options.
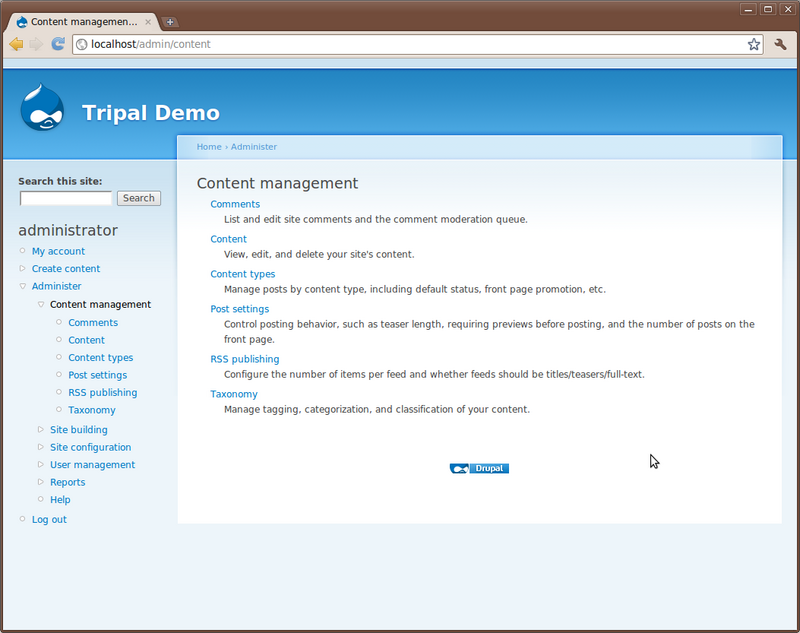
Click the Content link. The page shows all content available on the site. You will see the “About” and “Home” pages you created previously:
You’ll also notice a set of drop down boxes for filtering the content. For sites with many different content types and pages this helps to find content. You can use this list to click to view each page or to edit.
Site Building
Modules
Click the Site Building link on the let sidebar under the Administer link. You’ll see several new menu options: Blocks, Menus, Modules and Themes. First click Modules
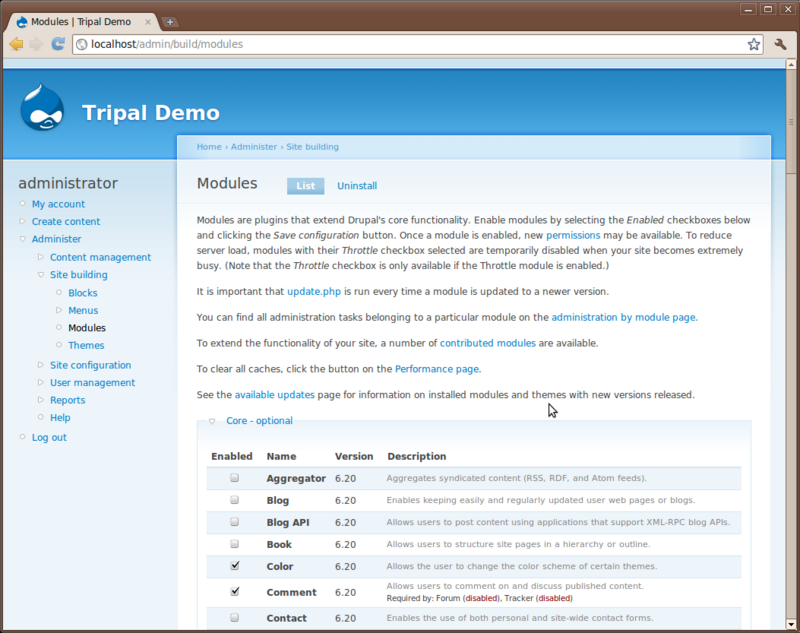
Here is where you will see the various modules that make up Drupal. Take a minute to scroll through the list of these and read some of the descriptions. The modules you see here are core modules that come with Drupal. Those that are checked come pre-enabled. Those that are not checked we will need to install. For this tutorial we will need two additional modules that are not yet installed. Locate the modules Path and Search and check the box next to each of those. Scroll to the bottom and click ‘Save configuration’.
The Path and Search modules are now installed. The Search module enables site-wide searching capabilities for our site and the Path module enables alternative naming of page URLs (we will discuss later).
Themes
Next, click the Themes link under Administer → Site Building on the left sidebar.
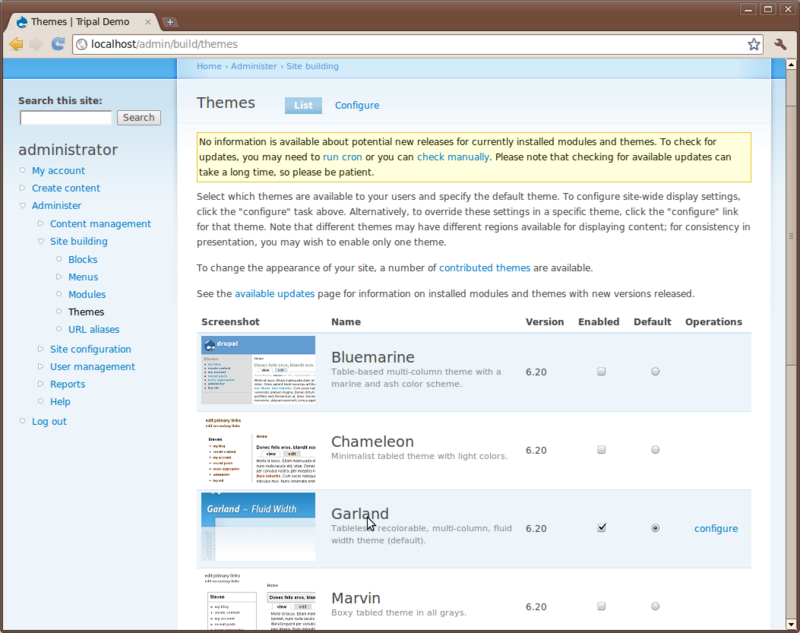
Here, you’ll see a list of themes that come with Drupal by default. If you scroll down you’ll see that one theme named Garland is enabled and set as default. The current look of the site is using the Garland them. Change the them by checking the Enable checkbox and the default radio button for the Pushbutton theme and then clicking Save configuration. Now you’ll see that the theme has changed.
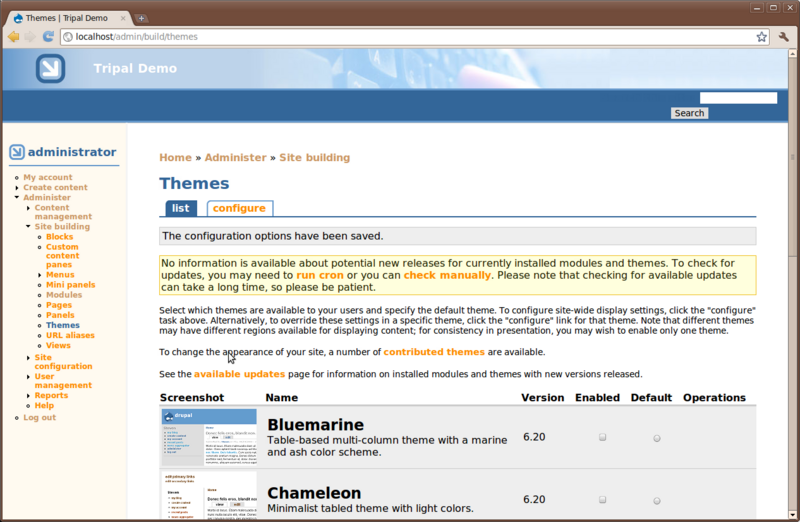
Blocks
Blocks in Drupal are used to provide content in regions of a Drupal theme. For example, navigate to Adminster → Site Building → Blocks.
You’ll see that regions of the theme have been identified. Within the Sky theme you can see the regions with dashed lines around them. Also, you’ll see a list of available blocks. You can select where blocks will appear by selecting the region in the drop down list. Blocks may also be hidden, if desired, by selecting <none> in the dropdown.
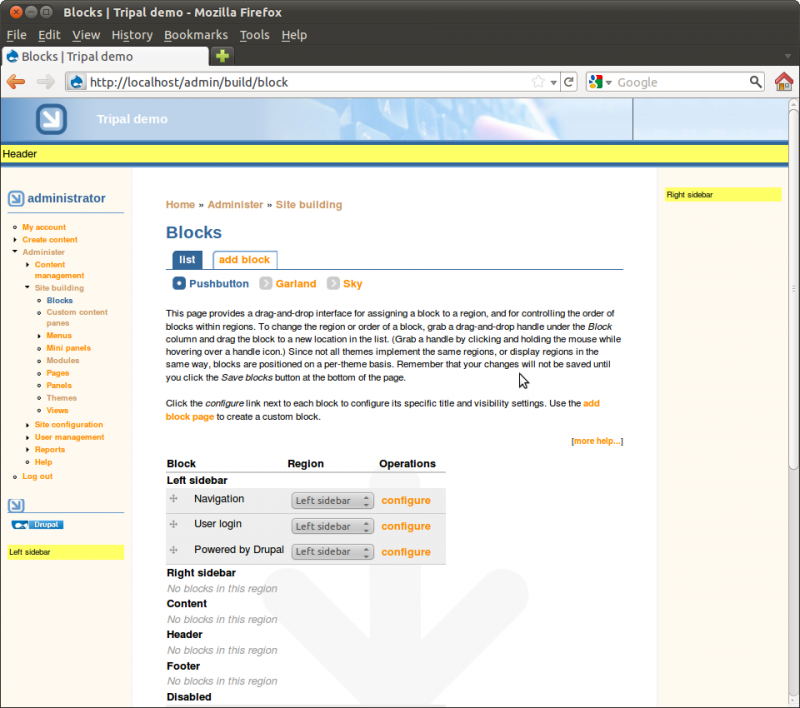
Take time to turn on and off blocks to see where they appear. Re-arrange blocks by dragging and dropping the cross-hairs beside each one. Be sure to leave the blocks in the configuration shown in the image below finished:
Menus
Drupal provides an interface for working with menus, including adding new menu items to an existing menu or for creating new menus. For the exercise in the Blocks section above we added the Primary links menu to the Content top section of the Sky theme. To view the Primary links menu, navigate to Administer → Site building → Menus.
Select the menu Primary links. You’ll see it currently has no item.
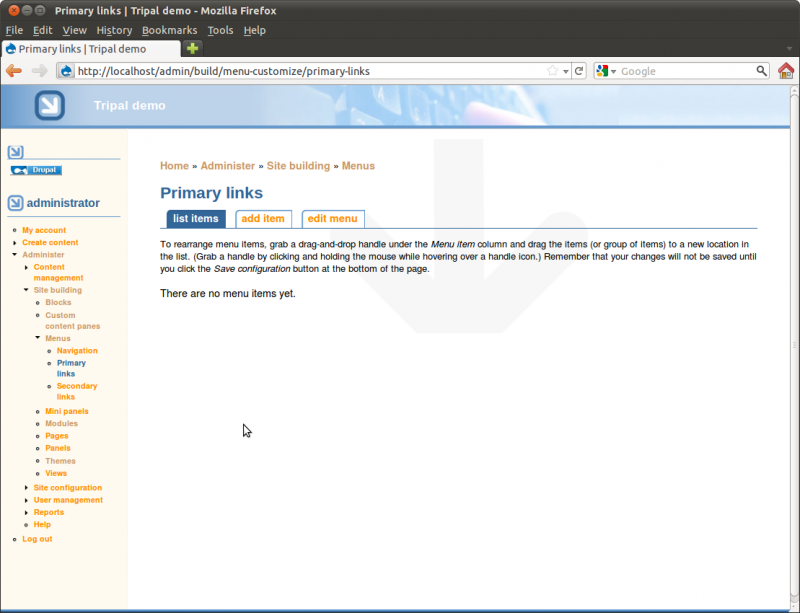
As a demonstration for working with menus we’ll add two menu items for the Home and About pages we created earlier. To do so, click the Add item tab. You will see a form for providing information about the menu item to be added.
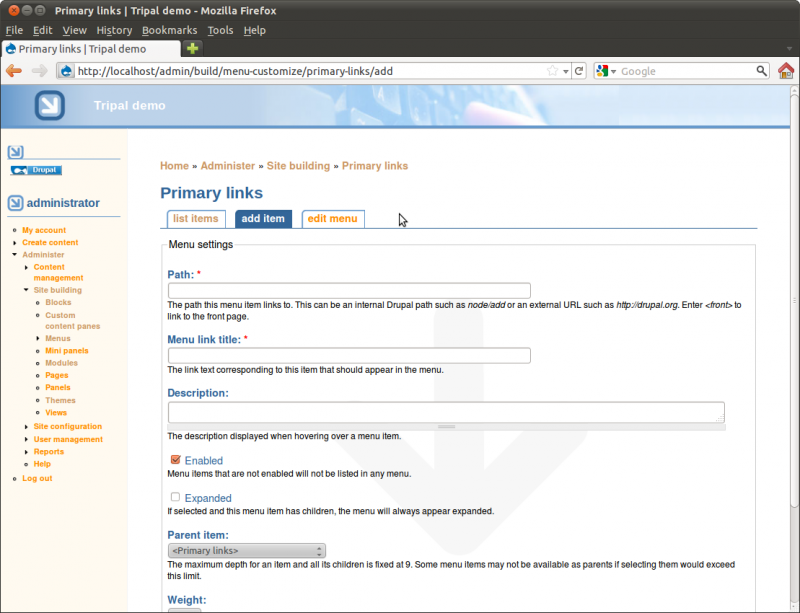
The first field is the path. We need to find the path for our home page.
The path for a page can be found in the address bar for the page. In Drupal pages of content are generally referred to as nodes. So, in the address bar for our home page you’ll see the address is http://localhost/node/1. Our about page should be http://localhost/node/2 (i.e the first and second pages we created).
The path for each of these nodes is simply node/1 and node/2.
Returning to our tab where we are adding a menu item, enter the path
node/1. We will set the fields in this ways:
- Path: node/1
- Menu Link Title: Home
- Description: Tripal Demo Home Page
- Enabled: checked
- Expanded: no check
- Parent item: <primary links>
- Weight: 0
The settings above will give the menu link a title of Home and put it on the Primary Links menu. We now have a Home menu item in the top just under the header, and our Home menu item now appears in the list of menu items for the Primary Links menu
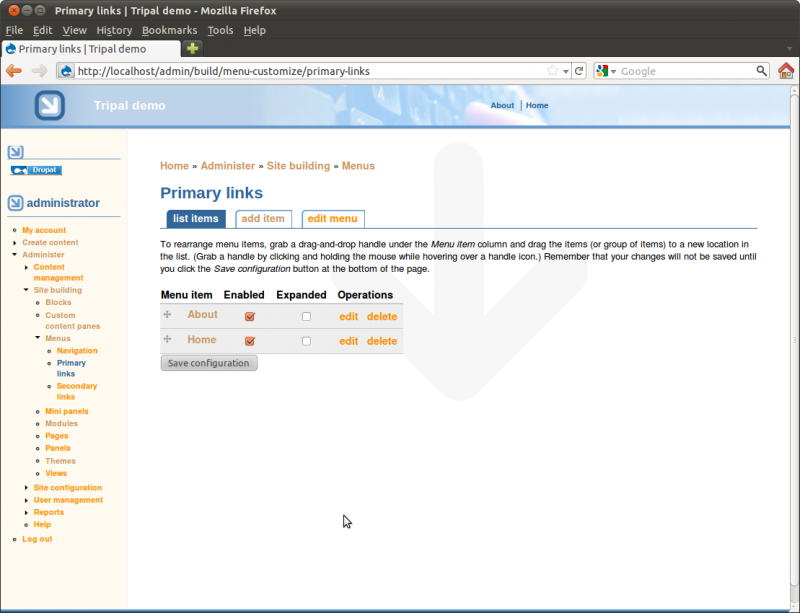
Using the insructions above to add a second menu item for our about page and arrange. Use the ‘weight’ value so that our Home link appears first and the About link appears second.
URL Path
As mentioned previously, the URL paths for our pages have node/1 and
node/2 in the address. This is not very intuitive for site visitors.
Earlier we enabled the Path module. This module will allow us to set
a more human-readable path for our pages.
To set a path, click on our new About page in the new menu link at the top and click the Edit tab. Scroll to the bottom of the edit page and you’ll see a section titled URL path setting. click to open this section.
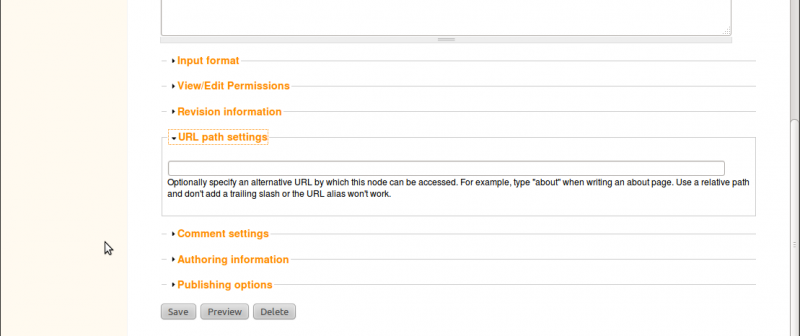
Since this is our about page, we simply want the URL to be http://localhost/about. To do this, just add the word about in the text box. You will now notice that the URL for this page is no longer http://localhost/node/2 but now http://localhost/about. Although, both links will still get you to our About page.
Now, use the instructions described above to set a path of ‘home’ for our home page.
Site Configuration
There are many options under the Administer → Site configuration page. Here we will only look at one of these at the moment–the Site Information page.
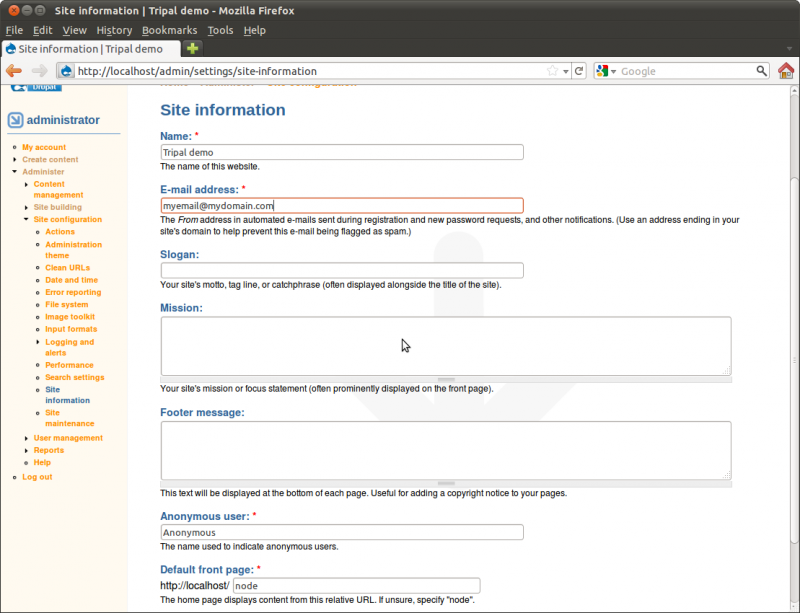
Here you will find the configuration options we set when installing the
site. You can change the site name, add a slogan, mission and footer
text to the. Towards the bottom there is a text box titled Default
front page. This is where we can tell Drupal to use our new Home
page we created as the first page visitors see when the view the site.
In this text box enter the text node/1. This is the address of our
home page. We must use the node number here and not our new URL path of
‘home’ that we just created. Let’s change the name of our site from
Tripal demo to My Community Genome Database and add a slogan:
Resources for Community Genomics.
Now, click the Save configuration button at the bottom. You’ll see our site name has changed at the top. Also, if we click the logo image at the top of the site and it will take you to the front page with our new home page appearing.
User Accounts
For this tutorial, we will not discuss in depth the user management infrastructure except to point out:
- User accounts can be created
- Users are assigned to various roles
- Permissions for those roles can be set to allow groups of users certain administrative rights or access to specific data.
Explore the Drupal User Management menu to see how users can be created, added to roles with specific permissions.
Prepare Drupal for Tripal
Theme Installation
Drupal allows us to install new themes. Installation of themes involves these steps:
- Locate and download a theme from the Drupal website (http://www.drupal.org/themes)
- Unpack the theme in the /var/www/sites/all/themes directory
- Return to the Drupal Administer → Site Building → Themes page and enable the theme
For this tutorial, we will use the Sky theme which is available from http://drupal.org/project/sky
cd /var/www/sites/all/themes
wget http://ftp.drupal.org/files/projects/sky-6.x-3.11.tar.gz
tar -zxvf sky-6.x-3.11.tar.gz
This should unpack the theme for us. Now, navigate to Administer → Site Building → Themes and enable the ‘Sky’ theme and set it as default:
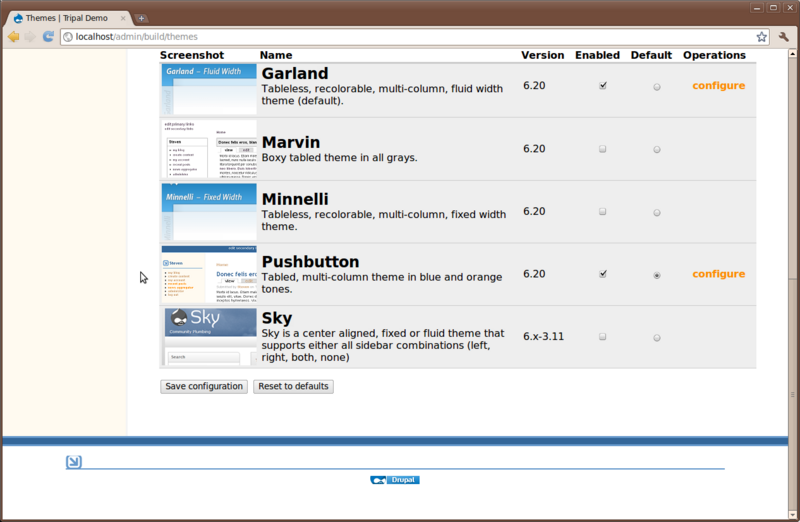
The sky theme was obtained at this address: http://drupal.org/project/sky
Theme Configuration
Here we return to theming. There are several configuration options that are available to help customize the theme for your site. These can be found by navigating to the Administer → Site Building → Themes page and clicking the Configure tab near the top.
Appearing under the Configure link will be small menu with a listing of every theme we have enabled. You should see the Sky theme at the end of this list. Click that theme because that is the one we are using and want to configure:
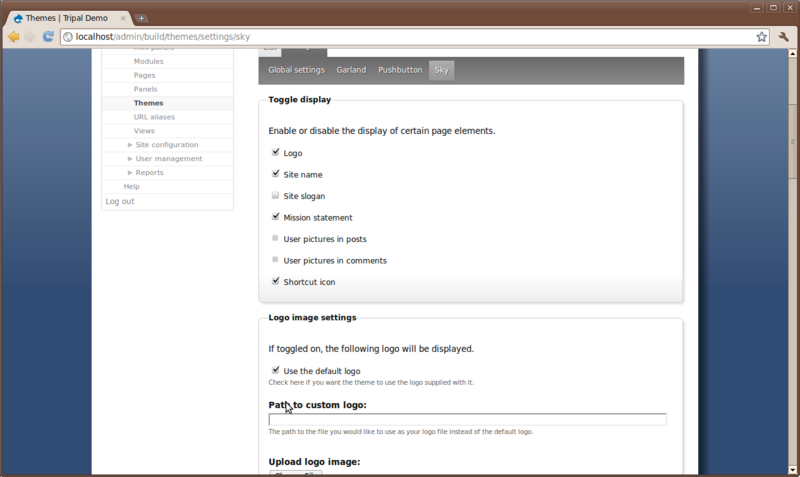
Here you can turn on and off the presence of the logo, site name, slogan, mission statement, etc. For this particular theme we can also adjust background colors and images, link colors, font style and size, and more. Notice when we added a slogan in a previous step but it did not appear anywhere on the site. To make it appear, check the box next to Slogan.
Also set the following for the theme:
- Set the ‘Custom Layout Width’ to be 1200px to give us more room
Then, click the Save Configuration button at the bottom. The pages inow a bit wider and our slogan appears.
3rd Party Modules
We can install new extension modules which we will need later. For this workshop we have several modules that we will need to install but which do not yet appear in the list of modules. To do this, we must follow these steps:
- Locate and download the extension modules from the Drupal website
- Unpack the module in the /var/www/sites/all/modules directory
- Check for a README.txt or INSTALL.txt for any further instructions for installation of the module
- Return the the Drupal ‘Administer’ -> ‘Site Building’ -> ‘Modules’ page and enable the module.
For an example, let’s install the Views module needed for this workshop. The Views module can be found here: http://drupal.org/project/views. We will download the current version as of the writing of this tutorial:
cd /var/www/sites/all/modules
wget http://ftp.drupal.org/files/projects/views-6.x-2.12.tar.gz
Now unpack the module:
tar -zxvf views-6.x-2.12.tar.gz
Check the README for additional installation instructions
cd views
ls
less README.txt
Use the space-bar to scroll through the README.txt file. Hit the ‘q’ key to quit
There are no other installation steps besides what we’ve done. So return to the Administer → Site Building → Modules page and enable the module.
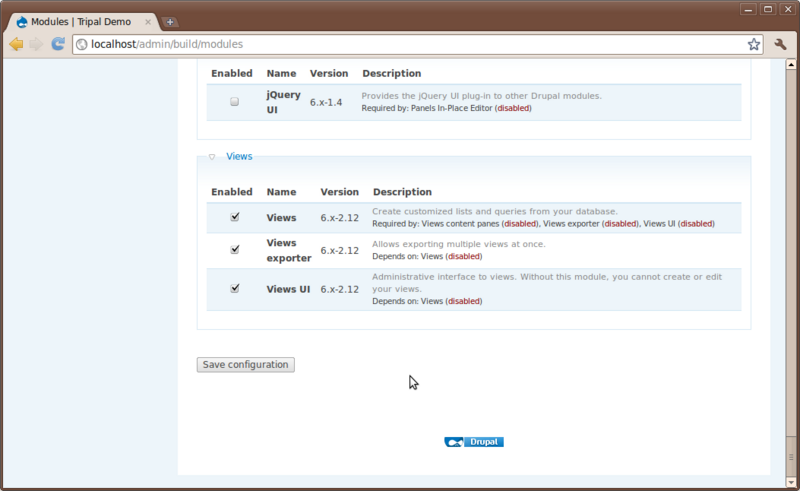
Notice that the Views package provided three different related modules and they all appear under a Views category.
for this Tutorial, CCK, Views, Views Data Export, and CKEditor should also be downloaded and installed following the same instructions above
cd /var/www/sites/all/modules
wget http://ftp.drupal.org/files/projects/views-6.x-2.16.tar.gz
wget http://ftp.drupal.org/files/projects/views_data_export-6.x-2.0-beta6.tar.gz
wget http://ftp.drupal.org/files/projects/cck-6.x-2.9.tar.gz
wget http://ftp.drupal.org/files/projects/ckeditor-6.x-1.11.tar.gz
For CKEditor the README file indicates we need to install the CKEditor package before we can enable this module. We must first get this package from online.
Here is a quick command for downloading this file
cd /var/www/sites/all/modules/ckeditor
wget http://download.cksource.com/CKEditor/CKEditor/CKEditor%203.6.4/ckeditor_3.6.4.zip
Now unzip the package and rename it according to the instructions
unzip ckeditor_3.6.4.zip
Now, return to our Drupal website and enable all the new modules.
For reference, the modules installed above can be found here:
- Views: http://drupal.org/project/views
- CCK: http://drupal.org/project/cck
- Views Data Export: http://drupal.org/project/views_data_export
- CKEditor: http://drupal.org/project/ckeditor
Drush
Drush is a command-line utility that allows for non-graphical access to the Drupal website. You can use it to automatically download and install themes and modules, clear the Drupal cache, upgrade the site and more. Tripal v1.0 supports Drush. For this tutorial we will use Drush and therefore we want the most recent version installed. Drush can be found on the Drupa website at http://drupal.org/project/drush.
To install drush first retrieve the most recent version from it’s Drupal project page. The current version at the writing of this document is version 7.x-5.7. While this version is intended for use with Drupal 7, it is backwards compatible with Drupal 6 and provides the most functionality.
We want Drush to reside in /usr/local which is where 3rd party software is normally installed. We’ll download the package to /usr/local/src and uncompress into /usr/local:
cd /usr/local/src
sudo wget http://ftp.drupal.org/files/projects/drush-7.x-5.7.tar.gz
cd /usr/local
sudo tar -zxvf src/drush-7.x-5.7.tar.gz
Next, we want the operating system to know about drush. We’ll create a symbolic link to the Drush executable in the /usr/local/bin directory where the operating systems looks for executables:
sudo ln -s /usr/local/drush/drush /usr/local/bin/drush
Finally Drush needs to perform updates the first time it is run, so we’ll run it with elevated privileges (using sudo) so that it can perform it’s updates. In the future we no longer need ‘sudo’ to run drush:
sudo drush
You must always run drush commands within the Drupal installation. It does not matter what subdirectory so long as you are in the Drupal directory sturcture. To see a list of available commands type the following:
cd /var/www/
drush
Tripal Installation
Get the Software
As of the writing of this Tutorial Tripoal v1.0 is near release. But until then is only available from the developmental source code. The source code is easily obtained using git. To download Tripal and the Extension modules change to the directory where Drupal keeps it’s modules:
cd /var/www/sites/all/modules
To obtain Tripal, issue the following ‘git commands:
git clone http://git.drupal.org/sandbox/spficklin/1337878.git tripal
cd tripal; git checkout 6.x-1.0-fix1; cd ../
We also want to obtain several Extension modules that will be used in this tutorial. Those modules are available on the Extensions Page of the Tripal website. However, these extension modules are also available via a git repository so we will use a git commands to obtain these.
git clone http://git.drupal.org/sandbox/spficklin/1578226.git tripal_blast_analysis
cd tripal_blast_analysis; git checkout 6.x-1.0-tripal_v1.0; cd ../
git clone http://git.drupal.org/sandbox/spficklin/1578234.git tripal_kegg_analysis
cd tripal_kegg_analysis; git checkout 6.x-1.0-tripal_v1.0-fix1; cd ../
git clone http://git.drupal.org/sandbox/spficklin/1578232.git tripal_interpro_analysis
cd tripal_interpro_analysis; git checkout 6.x-1.0-tripal_v1.0; cd ../
git clone http://git.drupal.org/sandbox/spficklin/1578230.git tripal_go_analysis
cd tripal_go_analysis; git checkout 6.x-1.0-tripal_v1.0; cd ../
git clone http://git.drupal.org/sandbox/spficklin/1578246.git tripal_unigene_analysis
cd tripal_unigene_analysis; git checkout 6.x-1.0-tripal_v1.0; cd ../
The above commands will download the main tripal package as well as the Blast, KEGG, InterPro, GO and Unigene extension modules. Tripal also has a theme as well. Change to the theme directory:
cd /var/www/sites/all/themes
And issue the following git commands:
git clone http://git.drupal.org/sandbox/spficklin/1342972.git tripal_theme
cd tripal_theme
git checkout 6.x-1.0
Installation
Previously in this Tutorial we enabled the Path and Search modules. The process for enabling the Tripal modules is the same. The site administrator can navigate to the Administer → Site Building → Modules page and enable each of the Tripal modules. However, Drush make it easier to enable modules from the command-line. To enable a module use the drush command in the following way:
drush pm-enable <module name>
First, we must enable the tripal_core module. Enter the following command
drush pm-enable tripal_core
Now that the core module is enabled, we must next install Chado. In the web browser, navigate to Administer → Tripal Management → Install Chado Schema. Since this is a fresh install, select the option to install Chado v1.2 and click the button Install/ugrapde Chado
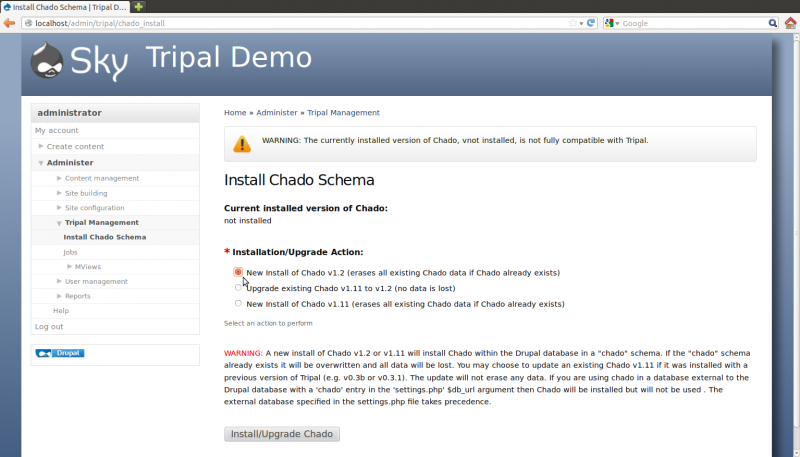
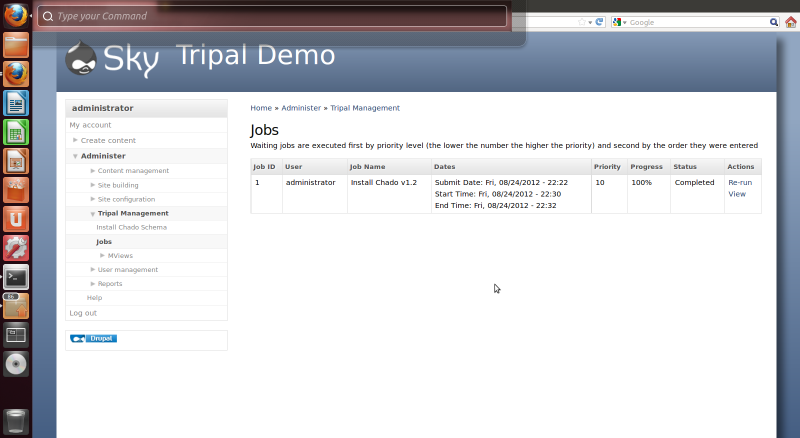
After the button is clicked a message will appear stating “Job ‘Install Chado v1.2’ submitted. Check the jobs page for status”. Click the jobs page link to see the job that was submitted:
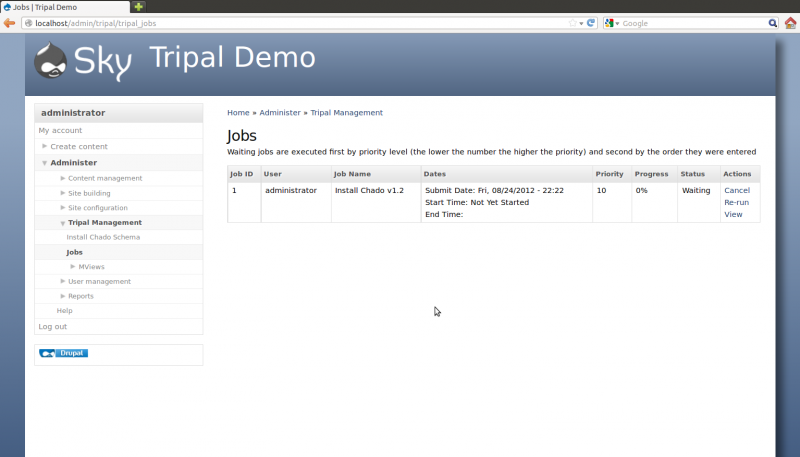
The job is waiting in the queue until the Tripal jobs system wakes and tries to run the job. The jobs management subsystem allows modules to submit long-running jobs, on behalf of site administrators or site visitors. Often, long running jobs can time out on the web server and fail to complete. The jobs system runs separately in the background using the command-line on an automated schedule but jobs are submitted through the web interface by users.
So, in the example above we now see a job for installing Chado. The job view page provides details such as the name of the job, The user who submitted the job, dates that the job was submitted and job status.
Jobs in the queue can be executed in two ways:
- Manually through a command-line call
- Using the UNIX cron to automatically launch the command-line
When we installed Drupal we installed a Cron job to allow the software to run housekeeping tasks on a regular bases. Tripal needs a cron entry as well to allow for regular execution of jobs in the queue. We will need to add a second cron entry:
sudo crontab -e
A word on text editors such as nano.
Add this line to the crontab
0,15,30,45 * * * * (cd /var/www; php ./sites/all/modules/tripal/tripal_core/tripal_launch_jobs.php administrator ) > /dev/null
This entry will run the Tripal cron every 15 minutes as the administrator user. For this tutorial we do not want to wait 15 minutes at the most to execute our jobs. So, we will run the jobs manually. Tripal supports Drush and therefore has it’s own commands. We can use drush to manually launch the job:
drush trpjob-run administrator
We should now see that Chado is being installed!
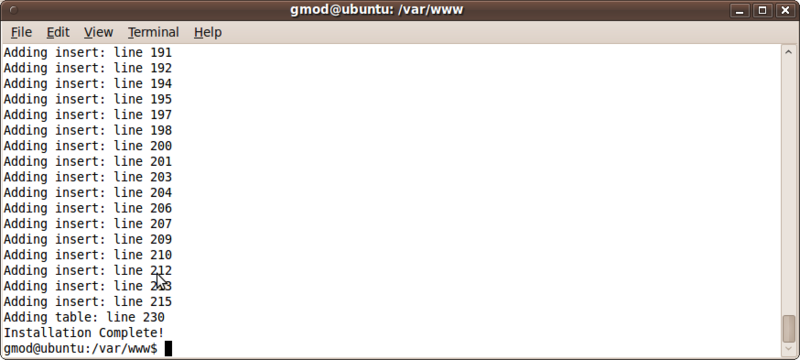
Also, we see that the job has completed when refreshing the jobs management page:
Now that Chado is installed, we can continue with installation of the remaining Tripal modules. These modules should be installed in the following order one at a time. If you install them all at once you may encounter errors later. Install the modules in the following way (and order):
drush pm-enable tripal_db
drush pm-enable tripal_cv
drush pm-enable tripal_organism
drush pm-enable tripal_analysis
drush pm-enable tripal_feature
Next, we have a few pre-requisite modules we must install. These are Views and Views Data Export modules. Previously in the tutorial these modules were installed, but to be thorough we show to install them here using drush.
First we need to patch the views_data_export module to work properly with Tripal. A patch is availabie inside of the tripal_views module. To apply the patches execute the following steps:
cd /var/www/sites/all/modules/views_data_export
patch -p1 < ../tripal/tripal_views/patches/views_data_export-postgresql-1293788-2.patch
Now enable the modules:
drush pm-enable views
drush pm-enable views_data_export
Now, enable the tripal_views modules and the remaining extension modules
drush pm-enable tripal_views
drush pm-enable tripal_analysis_blast
drush pm-enable tripal_analysis_go
drush pm-enable tripal_analysis_interpro
drush pm-enable tripal_analysis_kegg
There are more Tripal modules that can be enabled (e.g. tripal_project, tripal_stock, etc.). But for this tutorial we will only be using the modules we enabled above.
The Tripal modules create directories in the /var/www/sites/default/files directory. By default, Drupal expects the ‘sites/default/files’ directory to be writeable by the web server. Because we installed the Tripal mdoules using Drush we need to reset the permissions for the web user. Execute the following command to give the web user group permission to write to that directory
sudo chown -R ubuntu:www-data /var/www/sites/default/files
sudo chmod -R g+rw /var/www/sites/default/files
The last component we need to enable is the Tripal base theme. This theme provides the necessary look-and-feel to the data presented by Tripal. Installation is the same as for modules:
drush pm-enable tripal
The Tripal theme is not a full Drupal theme. It is intended to beincorporated into the site’s primary theme. In this tutorial we are currently using the sky theme. Therefore, we need to inform Drupal that the sky theme will be using Tripal as a base theme. To do this, change to the sky theme directory:
cd /var/www/sites/all/themes/sky
And edit the sky.info file
gedit sky.info
And add the following line to the bottom of the file:
base theme = tripal
If you do not wish to use the Sky theme, you simply need to find the corresponding .info file for your default theme and add the same line to the file.
Tripal is now installed!
Controlled Vocabularies: Installing CVs
Before we can proceed with populating our Chado table with genomic data we must first load some controlled vocabularies (i.e. ontologies). To do this, navigate to Administer → Tripal Management → Vocabularies. You’ll see a page describing the purpose of the module and how to use it. Click the link on the left sidebar titled ‘Load Ontology With OBO File’. You’ll see the following page:
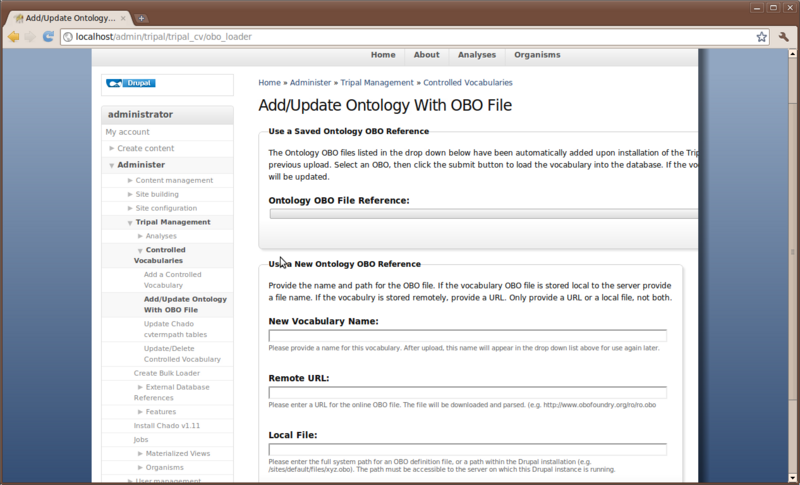
The Ontology loader will allow you to select a pre-defined ontology from the drop down list or allow you to provide your own to be loaded. If you provide your own, you give the remote URL of the OBO file or provide the full path on the local web server where the OBO file is located. In the case of a remote URL, Tripal first downloads and then parses the OBO file for loading. If you do provide your own OBO file it will appear in the saved drop down list for loading of future updates to the ontology.
For this tutorial, we need to install these ontologies:
- Chado feature properties
- Relationship ontology
- Sequence ontology
- Gene ontology.
Do so by selecting one and clicking the Submit button. Repeat this process for each of the three ontologies. You’ll notice each time that a job is added to the jobs subsystem.
Now manually launch these jobs
cd /var/www
drush trpjob-run administrator
Note: Loading the Gene Ontology will take several hours.
Setting Perimssions
Because we are logged on to the site as an admin user we are able to see all content. However, Drupal provides User Management infrastructure that allows the site admin to set which types of users can view the content on the site. By default there are two types of users anonymous and authenticated users. For this tutorial we want to set permissions so that anonymous visitors to the site can see the genomic content. To do this, navigate to Administer → user Management → Permissions. Here you will see permissions for all types of content.
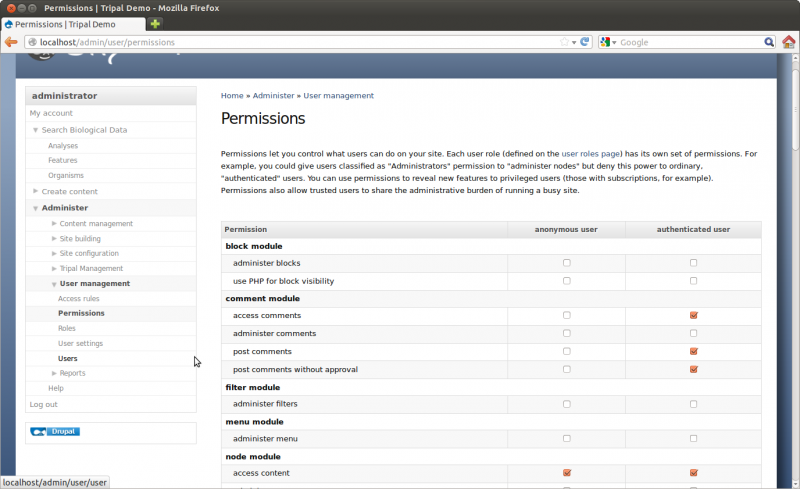
Scroll through the list of permissions and set the following for both anonymous and authenticated users:
- access chado_analysis content
- access chado_analysis_blast content
- access chado_analysis_go content
- access chado_analysis_interpro content
- access chado_analysis_kegg content
- access chado_cv content
- access chado_db content
- access chado_feature content
- access chado_organism content
Each time you install a new module you should always check the Permissions page and set any new permissions that may have been added by the new module.
Using Tripal
Creating Organism Pages
There are two ways to create pages for organism. If your organism is already in Chado then you can sync the organism. If it is not in Chado you will need manually create it using the Tripal web interface. The following two sections describe both methods.
What if Our Organism is Already in Chado?
Now that we have Chado loaded and populated we would like to create a home page for our species. Chado comes pre-loaded with a few species already, so we will check to see if our organism is already present. To do this navigate to Administer -> Tripal Management → Organisms → Configuration
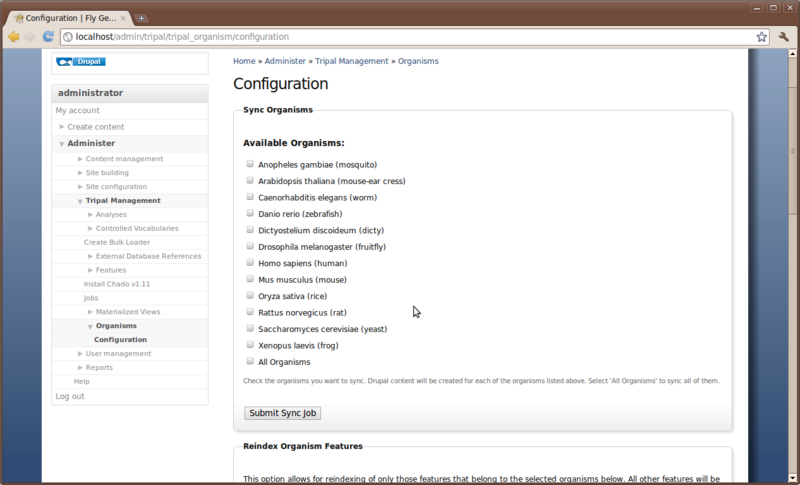
This configuration page has several different options. We will discuss two of these here. The first is the top section labeled Sync Organisms. In this section is a list of organisms. These are the organisms that come by default with Chado. If our organism is already in the list (e.g. Drosophila melenogaster) then we need to inform Drupal that we have data in Chado for which we would like a web page. This is what we call Syncing. We need to sync Drupal and Chado so that Drupal knows about our organism. To do this, click the check box next to Drosophila melenogaster and then click the Submit Sync Job.
As usual we want to run this job manually:
cd /var/www
drush trpjob-run administrator
Now that our organism is synced we should have a new page for Drosophila melenogaster. To find the page, click the Organisms menu item in the left side bar under Search Biological Data. This menu item was automatically added when we installed the Tripal Organism module. On this page we see a list of organisms that are present in Chado. Notice that only the fruitfly is clickable because it is the only organism synced.
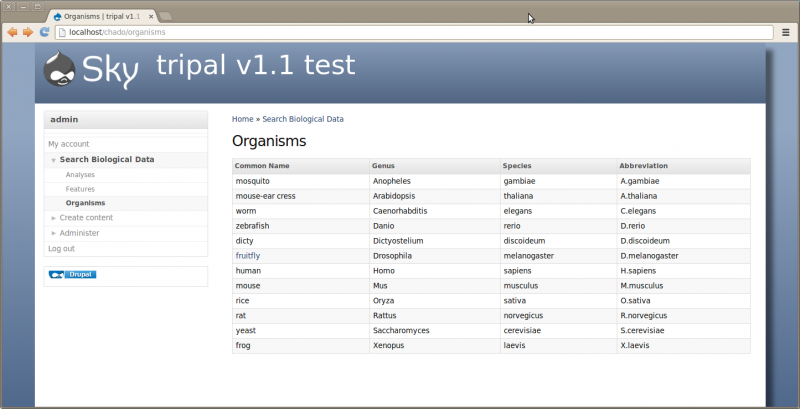
Now if we click the ‘fruitfly’ link it should take us to our new organism page:
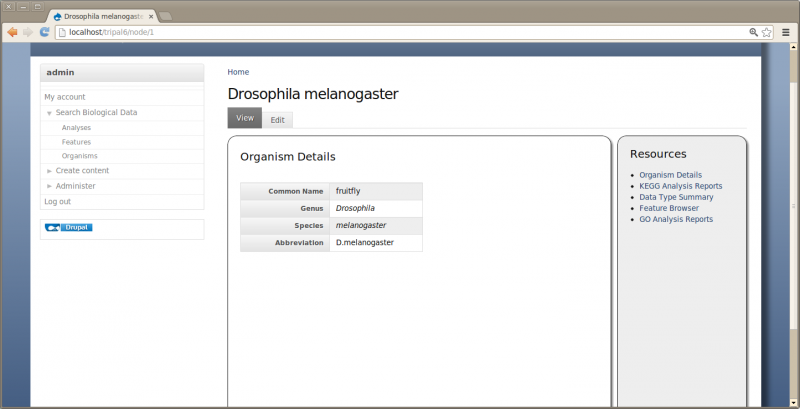
By default all Tripal pages have a center content section and a right side-bar section with links for Resources. However, this page is a bit empty. We need to add some details. Click the Edit tab towards the top of the page. Notice two if the fields are missing content: the description and the organism image
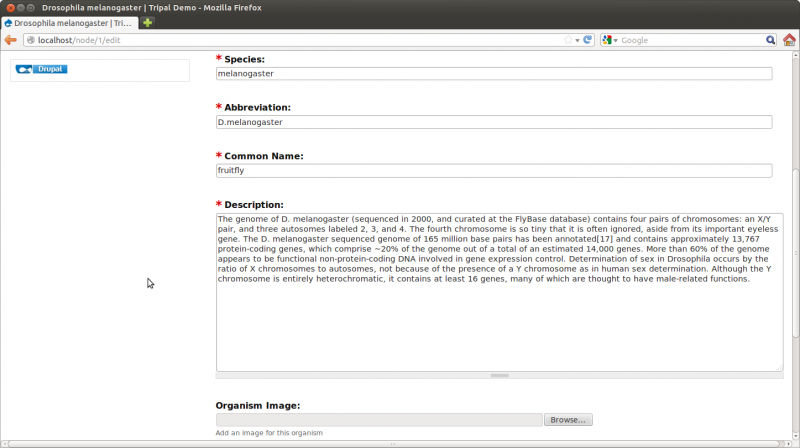
For the description add the following text (taken from wikipedia: http://en.wikipedia.org/wiki/Drosophila_melanogaster):
“The genome of D. melanogaster (sequenced in 2000, and curated at the FlyBase database) contains four pairs of chromosomes: an X/Y pair, and three autosomes labeled 2, 3, and 4. The fourth chromosome is so tiny that it is often ignored, aside from its important eyeless gene. The D. melanogaster sequenced genome of 165 million base pairs has been annotated[17] and contains approximately 13,767 protein-coding genes, which comprise ~20% of the genome out of a total of an estimated 14,000 genes. More than 60% of the genome appears to be functional non-protein-coding DNA involved in gene expression control. Determination of sex in Drosophila occurs by the ratio of X chromosomes to autosomes, not because of the presence of a Y chromosome as in human sex determination. Although the Y chromosome is entirely heterochromatic, it contains at least 16 genes, many of which are thought to have male-related functions.”
For the image, download this image below and upload it using the interface on the page.

Save the page. Now we have a more informative page:
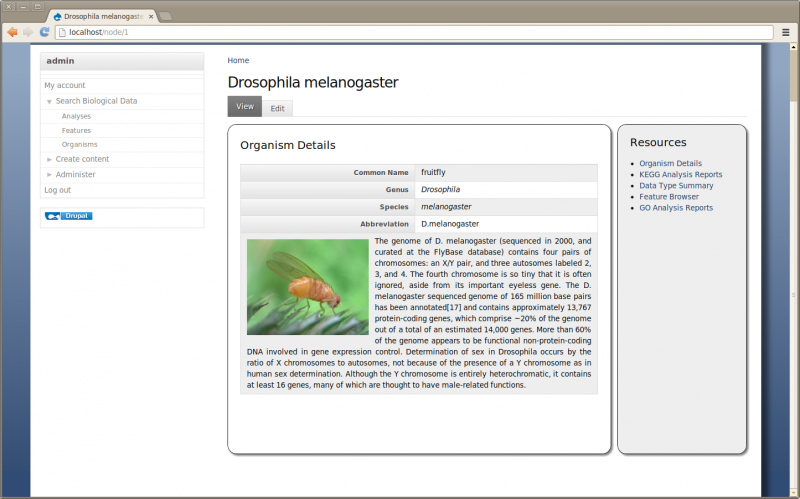
What if My Organism Is Not Present in Chado?
For this tutorial we will be loading data for Citrus sinensis (sweet orange), but this organism is not in Chado by default. We can easily add the organism using the Create Content page. You can find this link on the left side bar navigation menu. The Create Content page has many more content types than when we first saw it. Previously we only had Page and Story content types. Now we have more content types added by the Tripal Analysis, Organism, Feature and Extension modules .
To add a new organism simply click the Organism link and and fill in the fields with these values:
- Genus: Citrus
- Species: sinensis
- Abbreviation: C. sinensis
- Common name: Sweet orange
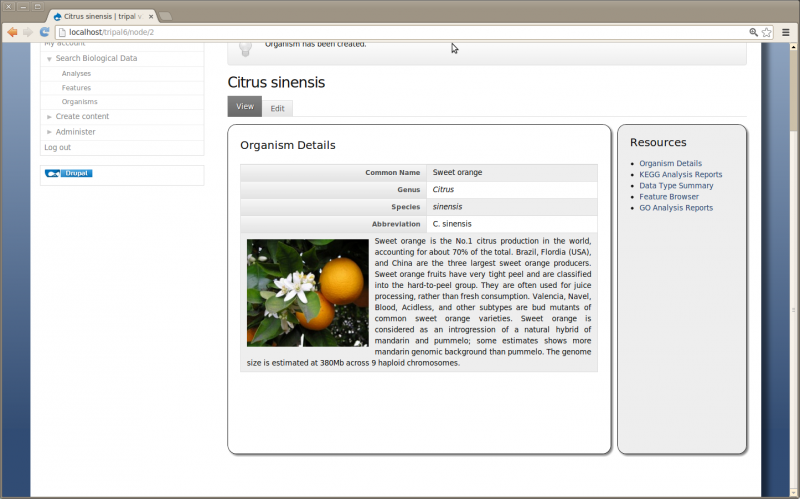
- Description: Sweet orange is the No.1 citrus production in the world, accounting for about 70% of the total. Brazil, Flordia (USA), and China are the three largest sweet orange producers. Sweet orange fruits have very tight peel and are classified into the hard-to-peel group. They are often used for juice processing, rather than fresh consumption. Valencia, Navel, Blood, Acidless, and other subtypes are bud mutants of common sweet orange varieties. Sweet orange is considered as an introgression of a natural hybrid of mandarin and pummelo; some estimates shows more mandarin genomic background than pummelo. The genome size is estimated at 380Mb across 9 haploid chromosomes.
And, use the following image:

Save the page and view the new Organism:
Creating an Analysis
For this tutorial, we will later import a set of genes, including associated mRNA, CDS, UTRs, etc. Tripal requires that an Analysis be associated with all imported features. This has several advantages, including:
- The source of features (sequences) can be traced. Even for feature simply downloaded from a database, it allows someone else to see where the features came from.
- Provides a mechanism for describing how the features were created (e.g. whole genome structural and functional annotation description)
- The analysis links all of the features together which can be useful for querying for specific features from an analysis.
To create an analysis for loading our genomic data, navigate to the Create contentand click on the link: Analysis
The analysis creation page will appear:
Here you can provide the necessary details to help others understand the source of your data. For this tutorial, enter the following:
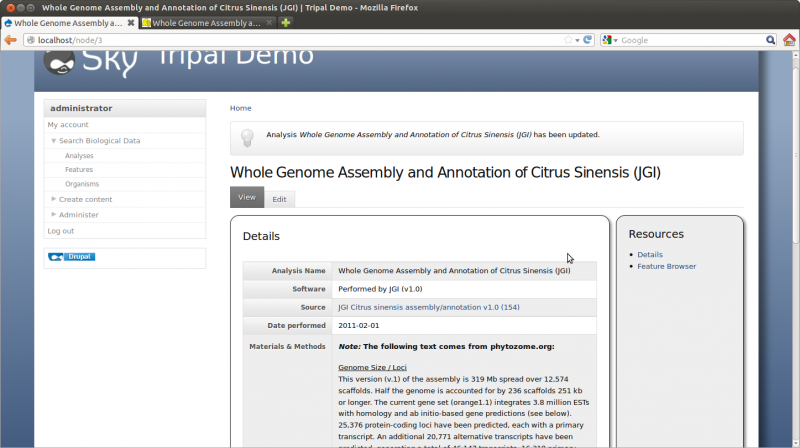
- Analysis Name: Whole Genome Assembly and Annotation of Citrus Sinensis (JGI)
- Program: Performed by JGI
- Program Version: v1.0
- Source Name: JGI Citrus sinensis assembly/annotation v1.0 (154)
- Source URI: http://www.phytozome.net/citrus.php
- Time Executed: Feb 1, 2011
- Description:
<p>
<strong><em>Note: </em>The following text comes from phytozome.org:</strong></p>
<p>
<u>Genome Size / Loci</u><br />
This version (v.1) of the assembly is 319 Mb spread over 12,574 scaffolds. Half the genome is accounted for by 236 scaffolds 251 kb or longer. The current gene set (orange1.1) integrates 3.8 million ESTs with homology and ab initio-based gene predictions (see below). 25,376 protein-coding loci have been predicted, each with a primary transcript. An additional 20,771 alternative transcripts have been predicted, generating a total of 46,147 transcripts. 16,318 primary transcripts have EST support over at least 50% of their length. Two-fifths of the primary transcripts (10,813) have EST support over 100% of their length.</p>
<p>
<u>Sequencing Method</u><br />
Genomic sequence was generated using a whole genome shotgun approach with 2Gb sequence coming from GS FLX Titanium; 2.4 Gb from FLX Standard; 440 Mb from Sanger paired-end libraries; 2.0 Gb from 454 paired-end libraries</p>
<p>
<u>Assembly Method</u><br />
The 25.5 million 454 reads and 623k Sanger sequence reads were generated by a collaborative effort by 454 Life Sciences, University of Florida and JGI. The assembly was generated by Brian Desany at 454 Life Sciences using the Newbler assembler.</p>
<p>
<u>Identification of Repeats</u><br />
A de novo repeat library was made by running RepeatModeler (Arian Smit, Robert Hubley) on the genome to produce a library of repeat sequences. Sequences with Pfam domains associated with non-TE functions were removed from the library of repeat sequences and the library was then used to mask 31% of the genome with RepeatMasker.</p>
<p>
<u>EST Alignments</u><br />
We aligned the sweet orange EST sequences using Brian Haas's PASA pipeline which aligns ESTs to the best place in the genome via gmap, then filters hits to ensure proper splice boundaries.</p>
Note:: Above we entered HTML. However, this is not the easiest way to enter text. When the ckeditor module is installed and properly setup the user is provided with editor tools which can appear like Microsoft Word 2003 controls. This makes it much easier to add text to any page. The setup for ckeditor is beyond the scope of this tutorial so we are required to enter HTML.
After saving, you should have the following analysis page:
Creating a Database Cross Reference
For our site, we want to create gene pages with sequences and have those pages link back to JGI where we obtained the genes. Therefore, we want to add a database reference for JGI. To add a new external databases, navigate to Administer → Tripal Management → Databases → Add External DB. The resulting page provides fields for adding a new database:
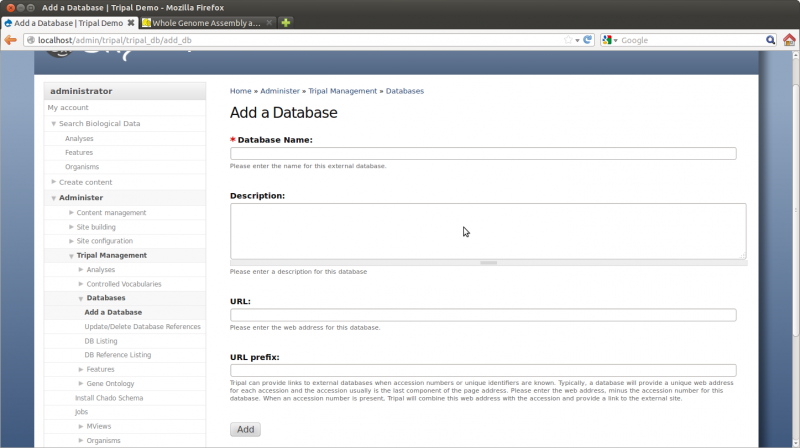
Enter the following values for the fields:
- Name: Phytozome
- Description: Phytozome is a joint project of the Department of Energy’s Joint Genome Institute and the Center for Integrative Genomics to facilitate comparative genomic studies amongst green plants
- URL: http://www.phytozome.net/
- URL prefix: http://www.phytozome.net/genePage.php?search=1&detail=1&er=1&crown&method=0&searchText=transcriptid%253A
The URL prefix is important as it will be used to create the links on our gene pages. Our gene name will be appended to this URL to create the link that will take us to the corresponding gene page on Flybase.
Click Add.
We now have added a new database!
Later we will also load Blast data. We need to create two new databases for those as well. Creat the following entries for NCBI nr, and ExPASy SwissProt:
- Name: NCBI nr
- Description: NCBI’s non-redundant database.
- URL: http://www.ncbi.nlm.nih.gov/
-
URL prefix: http://www.ncbi.nlm.nih.gov/protein/
- Name: ExPASy Swiss-Prot
- Description: A curated protein sequence database which strives to provide a high level of annotation, a minimal level of redundancy and high level of integration with other databases
- URL: http://expasy.org/sprot/
- URL prefix: http://www.uniprot.org/uniprot/
Loading Feature Data
Now that we have our organism and whole genome analysis ready, we can being loading genomic data. For this tutorial only a single gene from sweet orange will be loaded into the databsae. This is to ensure we can move through the tutorial rather quickly. The following datasets will be used for this tutorial:
- Citrus sinensis-orange1.1g015632m.g.gff3
- Citrus sinensis-scaffold00001.fasta
- Citrus sinensis-orange1.1g015632m.g.fasta
Download these to the /var/www/sites/default/files directory:
Loading a GFF3 File
The gene modules (e.g. gene, mRNA, 5_prime_UTRs, CDS 3_prime_UTRS) are stored in the GFF3 file downloaded in the previous step. We will load this GFF3 file and consequently load our gene feature into the database. Navigate to Administer → Tripal Management → Features → Import a GFF3 file.
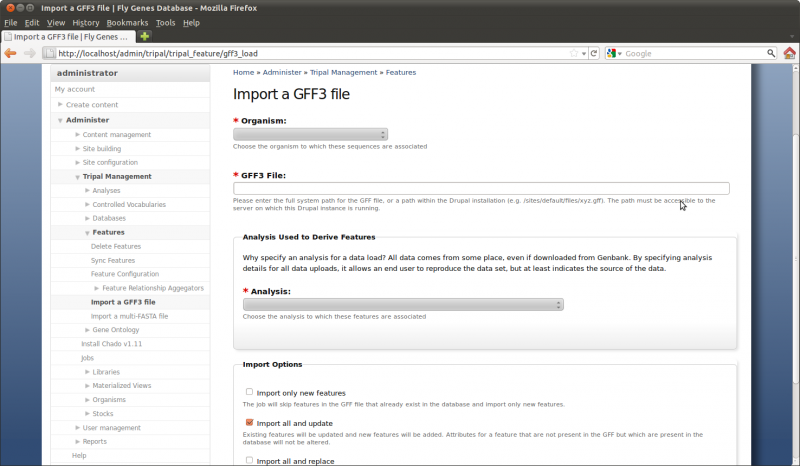
Perform the following:
- Choose the organism to which the GFF3 file belongs (in this case Citrus sinensis (sweet orange)
- Enter the path on the file system where our GFF file resides
(
/var/www/sites/default/files/Citrus sinensis-orange1.1g015632m.g.gff3) - Select the analysis named “Whole Genome Assembly and Annotation of Citrus sinensis…”.
- Select the check box “Import all and update”
Finally, click the Import GFF3 file button. You’ll notice a job was submitted to the jobs subsystem. Now, to complete the process we need the job to run. We’ll do this manually:
cd /var/www;
drush trpjob-run administrator
Note: For very large GFF files the loader can take quite a while to complete. If you find the loader is taking too long you can improve performance by using the Perl-based Chado loaders to load your GFF file.
A log file will be generated containing step-by-step output from loading the GFF file. The location of the log file is printed to the terminal window while the GFF file is loading. In the file you can look for any errors or problems that may have been encountred when loading your file. There should be no errors or warnings after loading the GFF file for this tutorial. A portion of the log file appears below:
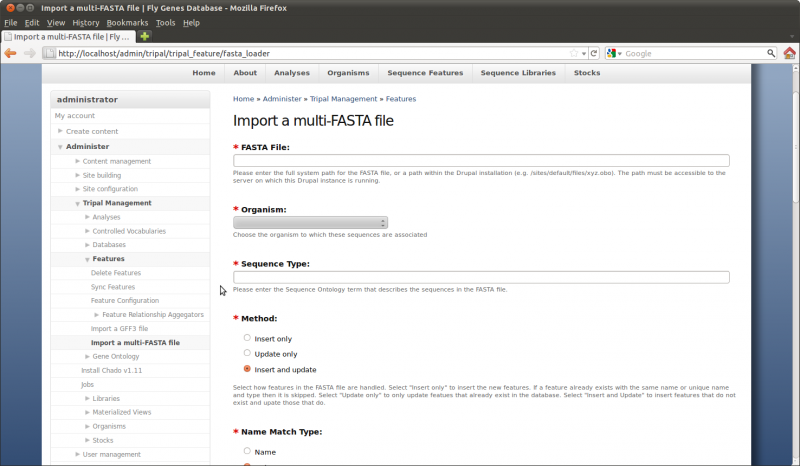
Loading FASTA files
Using the Tripal GFF loader we were able to populate the database with the genomic features for our organism. However, those features now need nucleotide sequence data. To do this, we will load the nucleotide sequences for the mRNA features and the scaffold sequence. Navigate to the Administer → Tripal Management → Features → Import a Multi-FASTA file Page
Before loading the FASTA file we must first know the Sequence Ontology (SO) term that describes the sequences we are about to upload. We can find the appropriate SO terms from our GFF file. In the GFF file we see the SO terms that correspond to our FASTA files are ‘scaffold’ and ‘mRNA’.
IMPORTANT: It is important to ensure prior to importing, that the FASTA loader will be able to appropriately match the sequence in the FASTA file with existing sequences in the database. Before loading FASTA files take special care to ensure the definition line of your FASTA file can uniquely identify the feature for the specific organism and sequence type. For example, in our GFF file an mRNA feature appears as follows:
scaffold00001 phytozome6 mRNA 4058460 4062210 . + . ID=PAC:18136217;Name=orange1.1g015632m;PACid=18136217;Parent=orange1.1g015632m.g
Note that for this mRNA feature the ID is PAC:18136217 and the name is orange1.1g015632m. In Chado, features always have a human readable name which does not need to be unique, and also a unique name which must be unique for the organism and SO type. In the GFF file, the ID becomes the unique name and the Name becomes the human readable name.
In our FASTA file the definition line for this mRNA is:
>orange1.1g015632m PAC:18136217 (mRNA) Citrus sinensis
By default Tripal will match the sequence in a FASTA file with the feature that matches the first word in the definition line. In this case the first word is orange1.1g015632m. As defined in the GFF file, the name and unique name are different for this mRNA. However, we can see that the first word in the definition line of the FASTA file is the name and the second is the unique name. Therefore, when we load the FASTA file we should specify that we are matching by the name because it appears first in the definition line.
If however, we cannot guarantee the that feature name is unique then we can use a regular expressions in the Advanced Options o tell Tripal where to find the name or unique name in the definition line of your FASTA file.
IMPORTANT: When loading FASTA files to update existing features, always choose “Update only” as the import method. Otherwise, Tripal may add the features in the FASTA file as new features if it cannot properly match them to existing features.
Now, enter the following values in the fields on the web form:
- FASTA file: /var/www/sites/default/files/citrus_sinensis-scaffold00001.fasta
- Organism: Citrus sinensis (Sweet orange)
- Sequence type: scaffold
- Method: Update only (we do not want to insert these are they should already be there)
- Name Match: Name
- Analysis: Whole Genome Assembly and Annotation of Citrus sinensis….
Click the Import Fasta File, and a job will be added to the jobs system. Run the job:
cd /var/www
drush trpjob-run administrator
Next do the same for the genes GFF:
- FASTA file: /var/www/sites/default/files/citrus_sinensis-orange1.1g015632m.g.fasta
- Organism: Citrus sinensis (Sweet orange)
- Sequence type: mRNA
- Method: Update only
- Name Match: Name
- Analysis: Whole Genome Assembly and Annotation of Citrus sinensis….
Now run this job:
cd /var/www;
drush trpjob-run administrator
Now the scaffold sequence and mRNA sequences are loaded
Note It is not necessary to load the mRNA sequences as those can be derived from the scaffold sequence and the alignment provided in the GFF file. However, in Chado the feature table has a ‘residues’ column and Tripal expects, in some cases, Tripal extension modules may expect the sequence to be present in that column (e.g. Tripal GO Analysis). Therefore it is best practice to load the sequence when possible.
The FASTA loader has some advanced options which we will not cover in this tutorial. But briefly, the advanced options allow you to create relationships between features and associate them with external databases. For example, the definition line for an mRNA is:
>orange1.1g015632m PAC:18136217 (mRNA) Citrus sinensis
Here we have more information that just the feature name. We have a ‘PAC’ ID which is a Phytozome accession number for that gene. We can therefore link this feature to Phyztome using External Database Reference section under the advanced options. We would need to provide the name of the database and regular expression to tell the loader where the accession number is found in the definition line. The database cross-reference is present in the GFF file and this assocation has already been made.
If the name of the gene to which this mRNA belonged was also on the definition line, we could use the Relationships section to link this mRNA with it’s gene parent. Fortunately, this information is in our GFF file and these relationships have already been made.
Creating Feature Pages
Now that we’ve loaded our feature data, we must “sync” them. Loading of the GFF file in the previous step has populated the feature tables of Chado for us, but now Drupal must know about these features. To sync features, navigating to Administer → Tripal Management → Features → Sync Features (unlike for syncing organisms, features have a separate page for syncing).
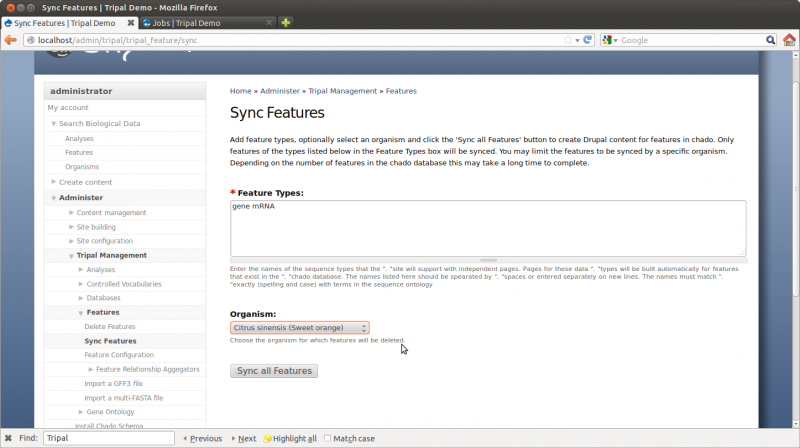
Here we can specify the types of features to sync and the organism. This allows us to create feature pages for different types of features for different organisms. Enter into the Feature Types the features that should have pages on the site. In this case, we want gene and mRNA pages. Features of these types were present in our GFF file.
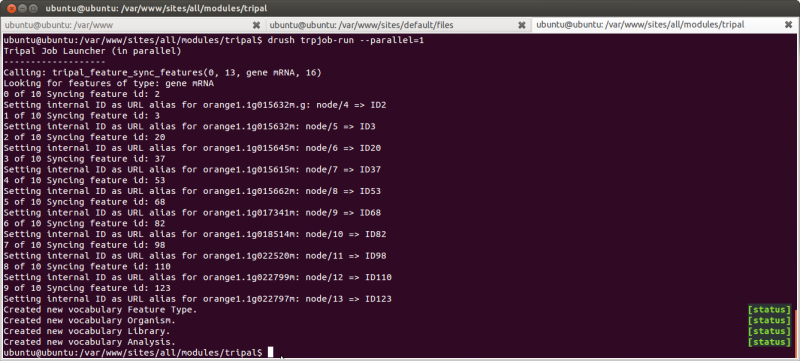
Next, select the organism “Citrus sinensis”, and click the “Sync all features” button. A job is then added to the jobs management system which we need to manually run rather than wait on the cron entry to run it.
cd /var/www
drush trpjob-run administrator
Our features are now synced:
Note: It is not necessary to sync all types of features in the GFF file. For example, do not sync the scaffold. The feature is large and would have many relationships to other features. By default, Tripal would try to show all of the relationships and alignments for the scaffold and the web page would time out. Tripal templates would need to be customized before a large feature is synced. Also, often GFF files have additional information such as blast, repeats, markers, etc. Only sync features that you know you will want pages for. For example, if you do not want a page for individual blast hits then do not sync match or match_part features.
Now, we can view our gene and mRNA pages. Navigate to Search Biological Data → Feautres. Click the Show button without adding any search criteria. The results will be all available features.
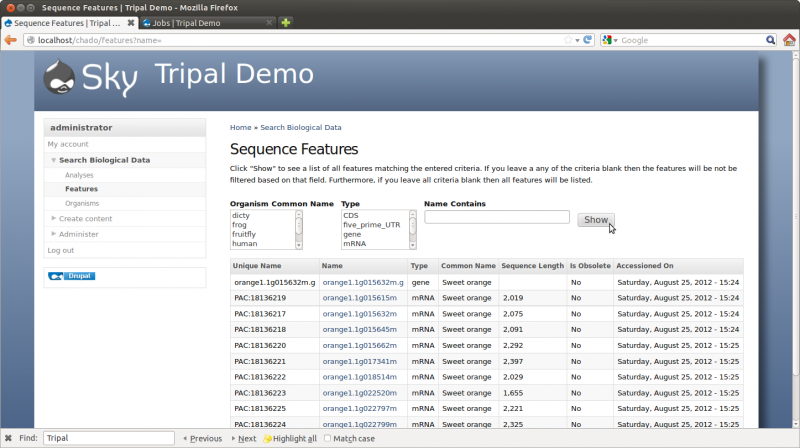
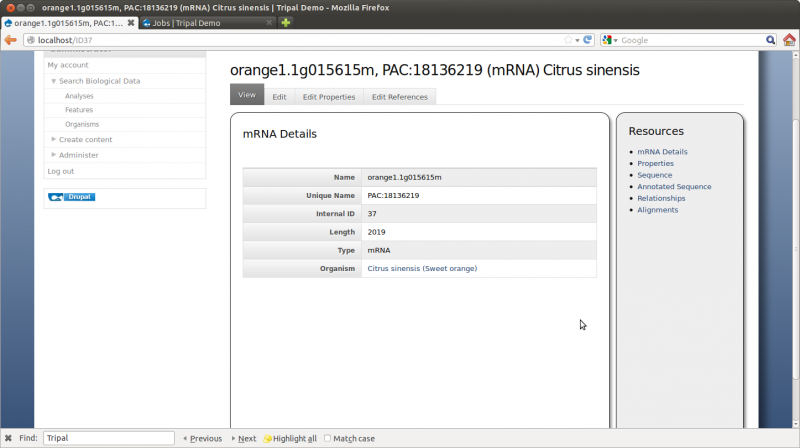
Here we can see the gene feature we added and its corresponding mRNA’s. Click on the mRNA named orange1.1g015615m. You can now see the details page with available resources on the side bar:
Materialized Views
Chado is very efficient as a data warehouse but queries can become slow depending on the number of table joins and amount of data. To help simplify and speed these queries, materialized views can be employed.
A materialized views will take an SQL query like this
SELECT DISTINCT CVT.name,CVT.cvterm_id, CV.cv_id, CV.name
FROM {cvterm_relationship} CVTR
INNER JOIN cvterm CVT on CVTR.object_id = CVT.cvterm_id
INNER JOIN CV on CV.cv_id = CVT.cv_id
WHERE CVTR.object_id not in
(SELECT subject_id FROM {cvterm_relationship})
And reduce it into this:
SELECT * FROM cv_root_mview WHERE cvterm_id = 100
For this to work a table named cv_root_mview is created and populated with the results of the larger query. The query on the materialized view is more simple and faster. A side effect, however is redundant data, with the materialized view becoming stale if not updated regularly.
Tripal provides a mechanism for populating and updating these materialized views. These can be found on the Administer → Tripal Management → Materialized Views page.
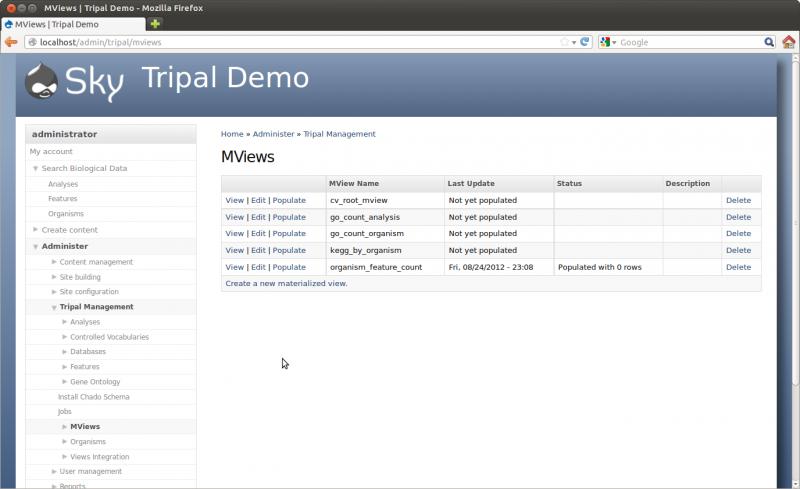
Here we see several materialized views. These were installed automatically by the various Tripal modules. To update these views, click the Populate button for each one.
So, let’s run these two jobs by clicking the Populate link next to each of the the view name. This will submit jobs to populate the views with data. Now, run the jobs:
cd /var/www
drush trpjob-run administrator
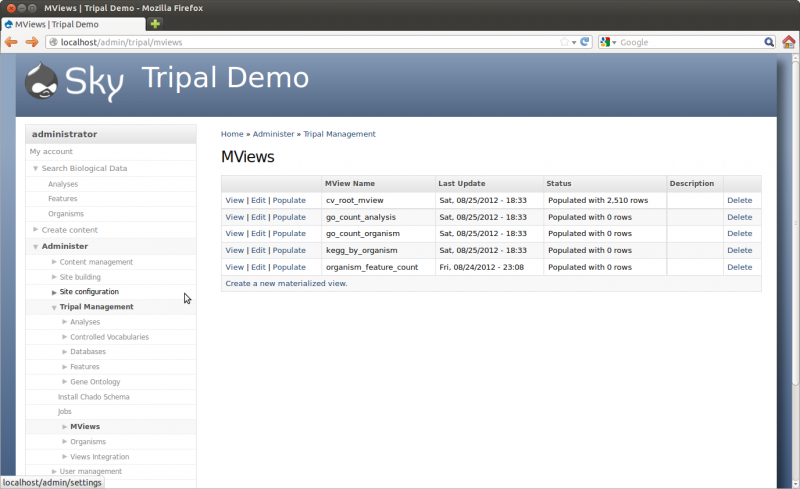
You can now see that all views are up-to-date on the Materialized Views Page. The number of rows in the view table is shown:
Feature Page Configuration
The feature configuration page allows us to perform configuration changes for the entire site. Navigate to the Administer → Tripal Management → Features → Feature Configuration page.
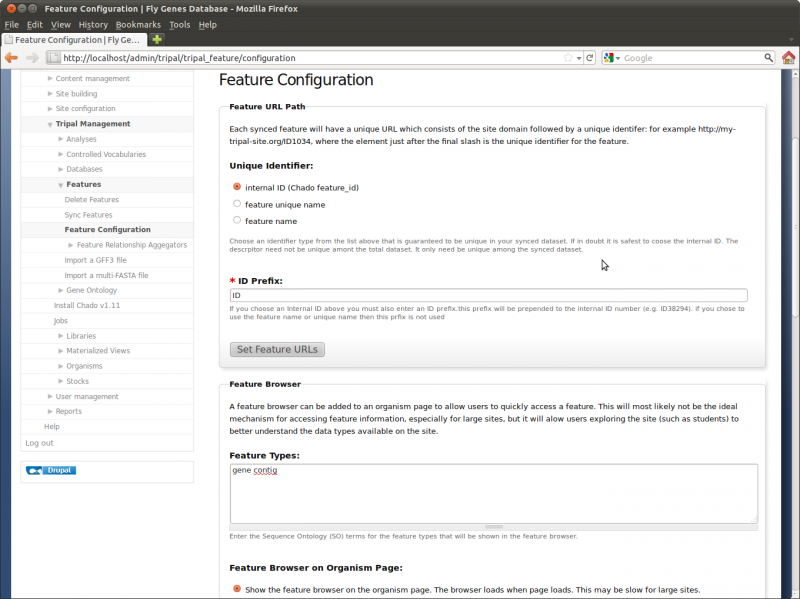
First, we can alter the feature URL path. This is the URL path that visitors can bookmark or link to for each feature. The URL would be http://localhost/[identifier] where [identifier] could be the name of the feature, unique name or internal ID number. If you are certain that all features can be uniquely identified by a name or unique name then select one of those, otherwise the internal ID is guaranteed to always be unique. If we choose to use an internal ID, we can specify a prefix for the internal ID number when it appears on the URL path. For example, if we leave the default prefix of ‘ID’ and have a feature ID number of 283942, the feature ID would appear as ‘ID283942’.
For this tutorial, we want to set the URL for all of the features we loaded previously. To do this, click the option feature name and then click the ‘Set Feature URLs’. This will add a job to the jobs subsytem. We want to execute this job manually:
cd /var/www
drush trpjob-run administrator
The URLs for feature are now similar to http://localhost/orange1.1g015632m. Before URLs were contained the default ID numbers.
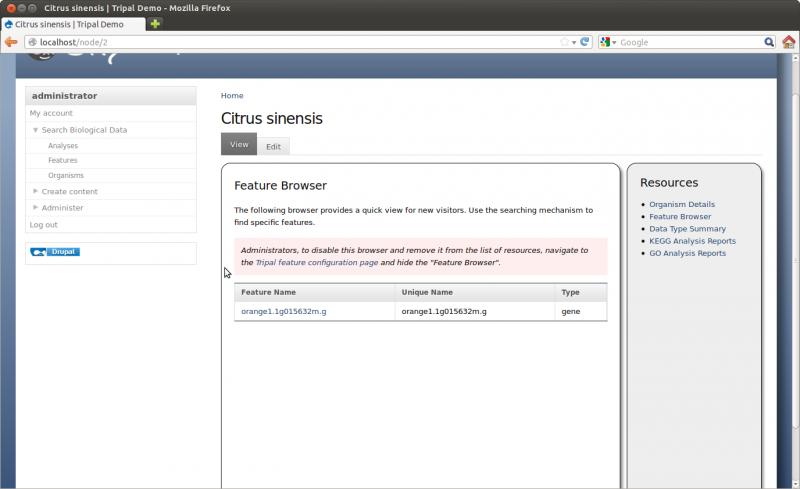
Next on the configuration page are Feature Browser settings. By default, Tripal will provide a browser on the organism page that allows a visitor to easily find a feature. For large sites with many features this would be an inefficient way to find a specific feature, but it does allow visitors who simply want to explore the site to quickly find example pages. This browser will only show synced features and will only show features of the type specified in the Feature Types box. We want to show ‘genes’ pages so alter the contents of this box to contain only the word ‘gene’. Optionally, you can turn this browser off by setting the appropriate radio buttons. If we then navigate to the organism page for Citrus sinensis and click the link on the right sidebar titled Feature Browser we can see the genes listed with links for each feature page.
Next on the configuration page is the Feature Summary setting. By default, on the organism page, Tripal will provide a list of all features associated with an organism and provide a pie-chart showing this list. You can turn this off by setting the appropriate radio button. You can also specify which feature types to show and can rename them to be more meaningful (e.g. reanme match to ‘BLAST match’). Below is a screen shot of the Date Type Summary available for the data we loaded. This is accessed by clicking on the link on the right side bar of the Citrus sinensis organism page.
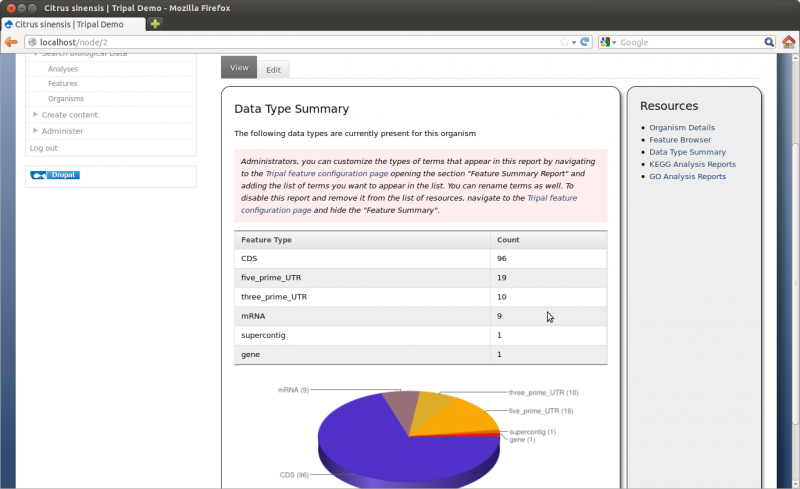
Note The data type summary is only available when the organism_feature_count materialized view is populated. Each time new data is added, this materialized view should be updated to capture the changes and have those shown in the summary.
Loading Functional Data Using Extension Modules
For this tutorial we will be loading functional data for our gene. To do this we will use the Blast, KEGG, and InterPro extension modules. These modules were installed previously. Blast, KEGG and InterPro analyses were completed prior to this tutorial and results files are avaialble for downloading:
- Citrus sinensis-orange1.1g015632m.g.iprscan.xml
- Citrus sinensis-orange1.1g015632m.g.KEGG.heir.tar.gz
- Blastx citrus sinensis-orange1.1g015632m.g.fasta.0 vs uniprot sprot.fasta.out
- Blastx citrus sinensis-orange1.1g015632m.g.fasta.0 vs nr.out
Download these files to the /var/www/sites/default/files directory.
Loading Blast Results
Configuring Blast Databases
Now that we have our features loaded we want to add some functional data as well. We need to create a new analysis page for our blast results. The Tripal Blast Analysis extension module will parse blast results and load them into Chado after a blast analysis page is created. However, before we create the page we need to ensure that the blast module can properly parse the blast hits. To do this, navigate to Administer → Tripal Management → Analyses””” → Blast Settings. The following page will appear.
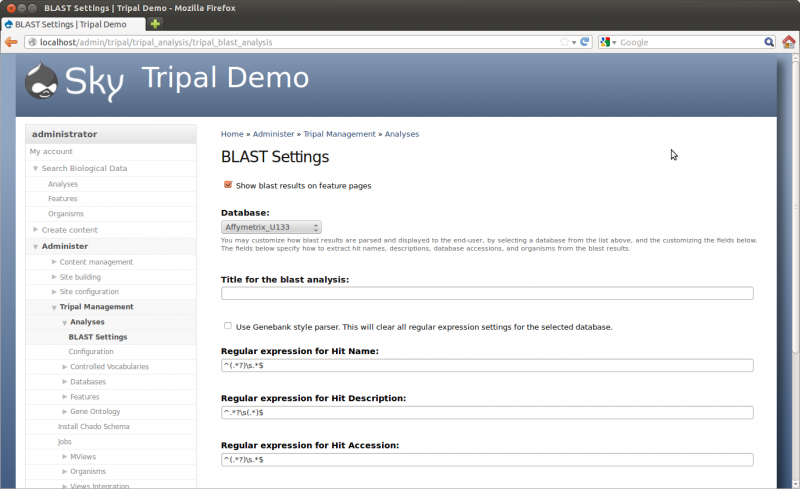
This page allows you to specify a different more meaningful name for the database to be displayed with blast results as well as to specify regular expressions for parsing blast htis. We will be adding blast results for the two databases we created earlier in the tutorial: ExPASy SwissProt and NCBI nr. First, select ExPASy SwissProt from the drop-down menu.
- Title for the blast analysis: (leave blank)
- Regular expression for Hit Name: ^sp\.*?\(.*?)\s.*?$
- Regular expression for Hit Description: ^sp\.*?\.*?\s(.*)$
- Regular expression for Hit Accession: ^sp\(.*?)\.*?\s.*?$
- Regular expression for Organism: ^.*?OS=(.*?)\s\w\w=.*$
- Organism Name: (leave blank)
Click Save Settings.
The regular expressions are designed to parse SwissProt-style hit names. For example, the following is an example of a match from SwissProt:
sp|P43288|KSG1_ARATH Shaggy-related protein kinase alpha OS=Arabidopsis thaliana GN=ASK1 PE=2 SV=3
Here the hit name is “KSG1_ARATH”. the accession is “P43288”, the hit description is “Shaggy-related protein kinase alpha OS=Arabidopsis thaliana” and the organism is “Arabidopsis thaliana”. The regular expressions are designed to extract these unique parts from the match text.
Note:: The match accession will be used for building web links to the external database. The accession will be appended to the URL Prefix set earlier when the database record was first created.
Now select the NCBI nr database from the drop-down and click the radio button Use Genebank style parser. This should disable all other fields.
Load the Blast Results
Now we can create out analysis page. Navigate to Create Content page and select the Analysis: Blast content type. Add the following values for this analysis. In the fields set the following values:
- Analysis Name: blastx Citrus sinensis v1.0 genes vs ExPASy SwissProt
- Program: blastall
- Program Version: 2.2.25
- Algorithm: blastx
- Source name: C. sinensis mRNA vs ExPASy SwissProt
- Time Executed: (today’s date)
- Materials & Methods: C. sinensis mRNA sequences were BLAST’ed against the ExPASy SwissProt protein database using a local installation of BLAST on in-house linux server. Expectation value was set at 1e-6
- Blast Settings
- Database: ExPASy SwissProt
- Blast XML File/Directory: /var/www/sites/default/files/blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_uniprot_sprot.fasta.out
- Blast XML File Extension: out
- Parameters: -p blastx -e 1e-6 -m 7
- Submit a job to parse the XML output: checked
- Keywords for custom search: checked
Click the Save button. You can now see our new Analysis.
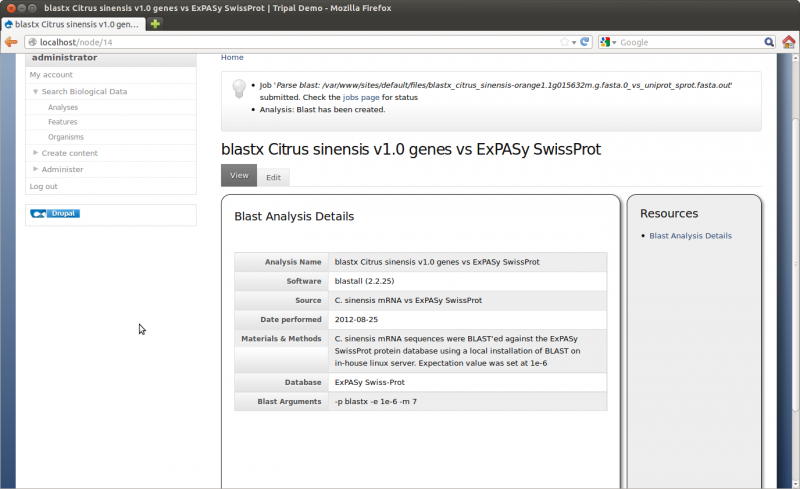
Now we need to manually run the job to parse the Blast results:
cd /var/www
drush trpjob-run administrator
The results should now be loaded. if we visit our feature page, for feature ‘orange1.1g015615m’ (http://localhost/orange1.1g015615m) we should now see blast results by clicking the ‘ExPASy SwissProt’ link on the right sidebar:
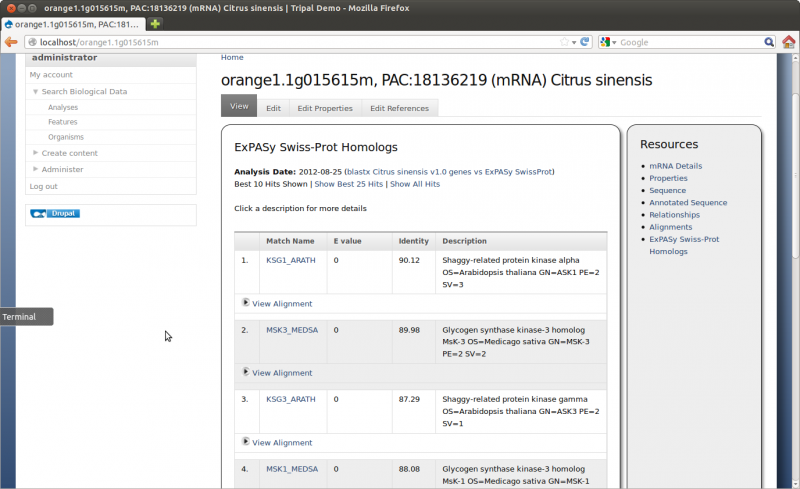
Now we want to add the results for NCBI nr. Repeat the steps above to add a new analysis with the following details:
- Analysis Name: blastx Citrus sinensis v1.0 genes vs NCBI nr
- Program: blastall
- Program Version: 2.2.25
- Algorithm: blastx
- Source name: C. sinensis mRNA vs NCBI nr
- Time Executed: (today’s date)
- Materials & Methods: C. sinensis mRNA sequences were BLAST’ed against the NCBI nr protein database using a local installation of BLAST on in-house linux server. Expectation value was set at 1e-6
- Blast Settings
- Database: NCBI nr
- Blast XML File/Directory: /var/www/sites/default/files/blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_nr.out
- Blast XML File Extension: out
- Parameters: -p blastx -e 1e-6 -m 7
- Submit a job to parse the XML output: checked
- Keywords for custom search: checked
Click the Save button and manually run the job:
cd /var/www
drush trpjob-run administrator
Return to the example feature page to view the newly added results: http://localhost/orange1.1g015615m
Loading InterProScan Results
Now we want to load results from an InterProScan. For this tutorial, these results were obtained by using a local installation of InterProScan installed on a computational cluster. However, you may choose to use Blast2GO or the online InterProScan utility. Results should be saved in XML format.
To create an analysis, navigate to the Create Content page and select the content type Analysis: Interpro. Add the following values for this analysis
- Analysis Name: InterPro Annotations of C. sinensis v1.0
- Program: InterProScan
- Program Version: 4.8
- Algorithm: iprscan
- Source name: C. sinensis v1.0 mRNA
- Time Executed: (today’s date)
- Materials & Methods: C. sinensis mRNA sequences were mapped to IPR domains and GO terms using a local installation of InterProScan executed on a computational cluster. InterProScan date files used were MATCH_DATA_v32, DATA_v32.0 and PTHR_DATA v31.0.
- InterPro Settings
- InterProScan XML File/Directory: /var/www/sites/default/files/citrus_sinensis-orange1.1g015632m.g.iprscan.xml
- Check the box ‘Submit a job to parse the Interpro XML output’
- Chekc the box ‘Load GO terms’
- Parameters: iprscan -cli -goterms -ipr -format xml
Click the Save button. You can now see our new Analysis.
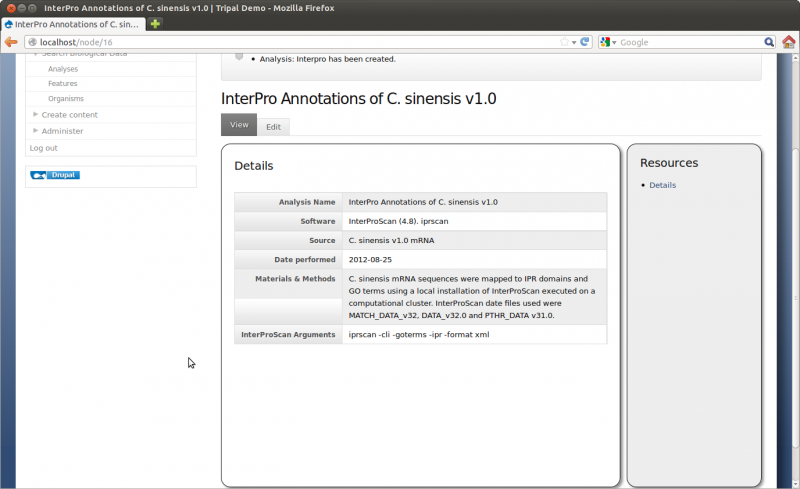
Now we need to manually run the job to parse the Inetpro results:
cd /var/www
drush trpjob-run administrator
The results should now be loaded. if we visit our feature page, http://localhost/orange1.1g015615m, we should now see interpro results by clicking on the “Interpro Report” link on the right sidebar.
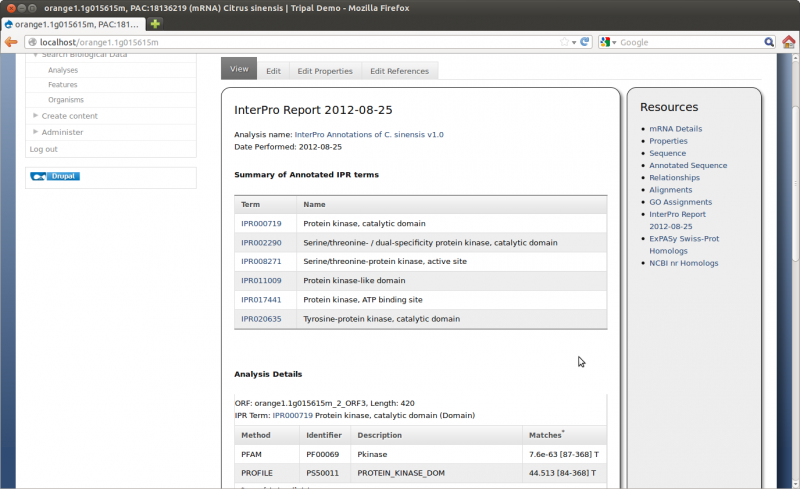
Viewing GO Terms
When we setup the InterPro analysis we requested that it parse GO terms from the InterProScan results. As a result, we now have a new GO Assigments item in the Resources sidebar. For our example feature (http://localhost/orange1.1g015615m), the results are as follows:
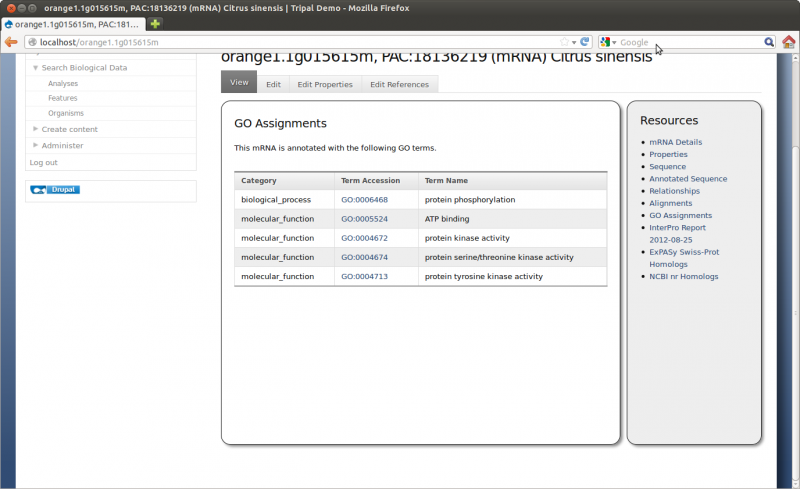
Because we now have GO terms associated with features we can setup the GO report that appears on the organism page. Navigate to the Citrus sinensis organism page and click the Go Analysis Reports in the Resources sidebar. A page appears with instructions to the site administrator that gives details for make the report visible.
Follow the instructions as presented on the page. Briefly, you need to
- Set the CV term paths for the three GO vocabularies
- Populate the go_count_analysis materialized view.
When complete the following report will be visible:
The GO report provides pie charts and an expandable tree for browsing results. Clicking on a GO term in the true will cause a box to appear with details about the term and a link to download a FASTA file of all featurs annotated with the term.
Loading KEGG Analysis Results
Now we want to load results from a KEGG/KAAS analysis (http://www.genome.ad.jp/tools/kaas/). The KAAS server receives as input a FASTA file of sequences and annotates those with KEGG terms. The tool also generates an heirarchy (heir) output file. This output file can be read directly by the Tripal Analysis KEGG module.
To create an analysis, navigate to the Create Content page and select the content type Analysis: KEGG. Add the following values for this analysis
- Analysis Name: KEGG analysis of C. sinensis v1.0
- Program: KEGG Automatic Annotation Server (KAAS)
- Program Version: 1.64a
- Source name: C. sinensis v1.0 genes
- Time Executed: (todays date)
- Materials & Methods: C. sinensis mRNA sequences were uploaded to the KEGG Automatic Annotation Server (KAAS) where they were mapped to KEGG pathways and orthologs. The SBH (single-directional best hit) was used with the genes data set being the defaults for genes.
- KEGG Settings
- KAAS hier.tar.gz Output File: /var/www/sites/default/files/citrus_sinensis-orange1.1g015632m.g.KEGG.heir.tar.gz
- Check the box “Submit a job to parse the kegg output into Chado”
Click the Save button. You can now see our new Analysis.
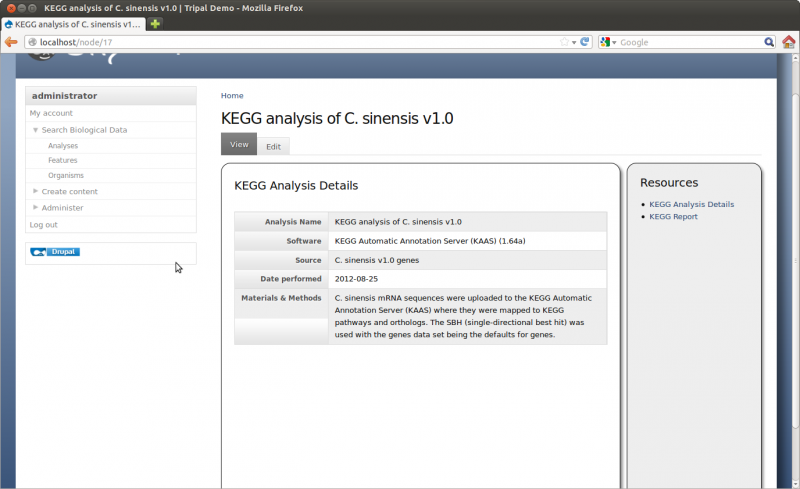
Now we need to manually run the job to parse the KEGG results:
cd /var/www;
drush trpjob-run administrator
Note: currently in the development version of Tripal there is a bug where the KEGG data is not loaded when the job runs. You must run the job a second time for the KEGG results to be loaded. To do this, edit the analysis by clicking the Edit tab at the top of the analysis, re-check the box titled “Submit a job to parse the kegg output into Chado”, click Save and then re-run the job.
If we navigate to our feature page (http://localhost/orange1.1g015615m) we can now see the KEGG results by clicking the KEGG Assignments link in the Resources sidebar.
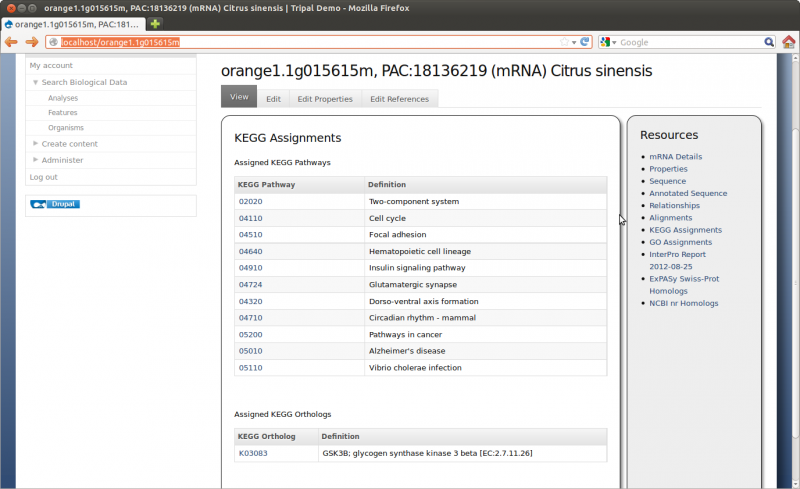
Simliar to the GO report. A KEGG report is also avilable on the analysis and the organism page. Navigate to the Citrus sinensis organism page and click the KEGG Analysis Reports in the Resources sidebar. A page with instructions is visible:
Follow the instructions on the page. Because we have already loaded the data we only need to popluate the kegg_by_organism materialized view. After populating the view we can now return to the organism page and view the KEGG report:
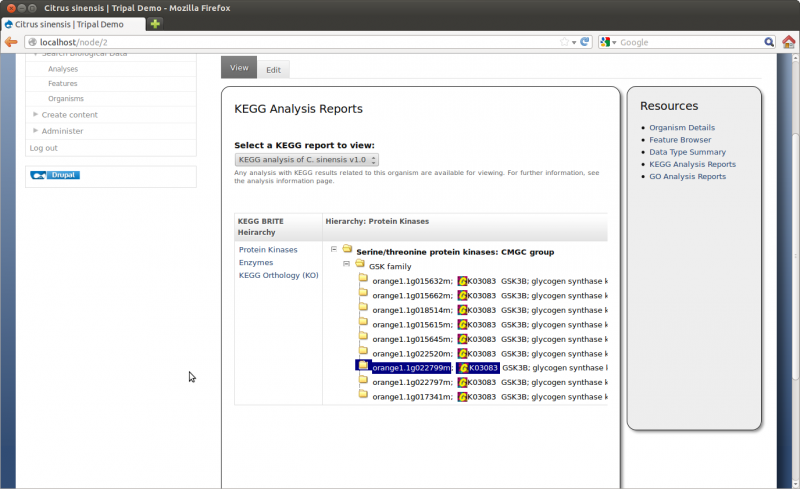
Site visitors can browse KEGG results by expanding the trees correspoding to the the Brite terms.
Customizing The Look-and-Feel of Tripal
The default look-and-feel of data presented by Tripal is set in Drupal-style template files. These template files can be found inside of the Tripal theme and theme folder of the Tripal Extension modules. Drupal allows you to customize the templates. For this tutorial we will not cover customization of template files. However, a tutorial for customizing the look-and-feel of the site using templates can be found in the Developers Handbook.
Adding Additional Resources
As mentioned previously, you can alter the look-and-feel of a Tripal site by customizing the templates. However, in the case of adding new items to the Resources sidebar of an organism, feature or analysis page, you can do this using the Content Construction Kit (CCK). The CCK is a 3rd-parth Drupal module that allows new fields to be added to any content. Using the CCK we will add new fields to our organism page which Tripal will recognize as new resources.
First, if not already installed, we must install the CCK module. We can do so easily using drush:
drush pm-enable content
drush pm-enable text
drush pm-enable number
Now, navigate to Administer → Content Types. The following page appears:
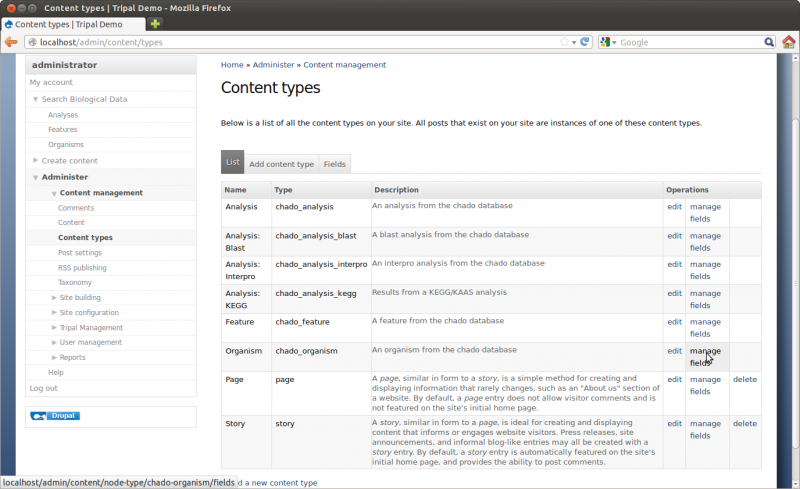
Beside the Organism content type, click on the link manage fields. The field editor page appears:
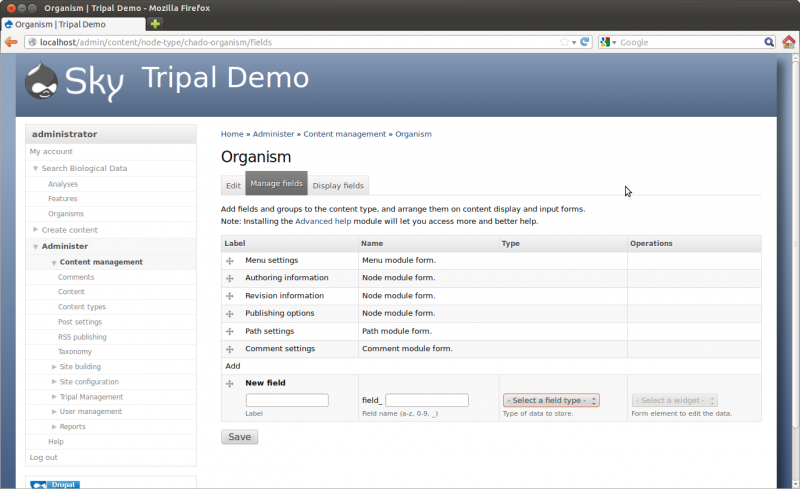
We need to add three specific fields. First adding the following values to the form:
Field #1
- Label: Resource Titles
- Field: resource_titles
- Type of data to store: Text
- Form element to edit the data: Text Field
Click Save button and the following field setup page appears:
File:Tripal-CCK-FieldSetup.png
On this page, leave all fields as default except one. Set the number of values to Unlimited and click the Save field settings button. We can now see the new field:
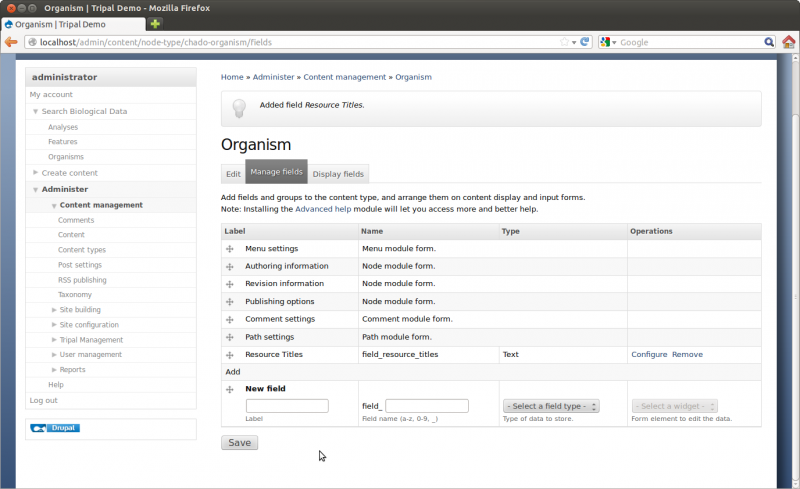
Finally, we don’t want this field to show up on the page but rather we want Tripal to handle it. If Tripal sees a field with the name ‘field_resource_titles’ it will automatically add it to the Resources sidebar and hence we do not need Drupal to display it. To exclude the field, click the Display fields tab. On this page there are two checkboxes, each under a column titled ‘Exclude’. Check both checkboxes and then click the Save button.

This new CCK field we created will let the site admin add new titles to the Resources sidebar. However, we now need content for any titles. We also may simply want to put links on the sidebar. We need to create two new CCK elements. Follow the same steps as described above to create the following fields:
Field #2
- Label: Resource Blocks
- Field: resource_blocks
- Type of data to store: Text
- Form element to edit the data: Text area (multiple rows)
- Settings
- Number of values: Unlimited
- Text processing: Filtered text (user selects input format)
Note that for the resource block a new setting is set for text processing. We want to allow HTML input.
Field #3
- Label: Resource Links
- Field: resource_links
- Type of data to store: Text
- Form element to edit the data: Text Field
- Settings
- Number of values: Unlimited
Don’t forget to exclude these two fields as well!
In summary, we created three new fields that will appear on our organims page. Tripal will recognize these new field names (e.g. field_resource_titles, field_resources_blocks and field_resource_links) and will automatically put new items on the Resources sidebar for titles and links. The resource blocks are then the text content that corresponds to the titles.
Now, we can return to our Citrus sinensis organism page, and click the ‘Edit’ tab at the top. Scroll to the bottom of the page and you will see our newly added fields.
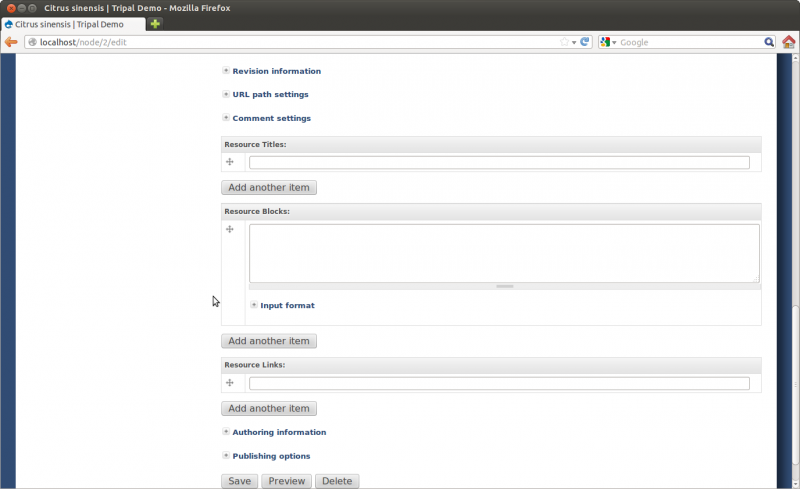
We will use these fields to add a link to the Phytozome page for Citrus sinensis and also a Downloads block that will allow the site visitor to download all of the data files we used in this tutorial. First, add the following link to the Resource Links text box:
C sinensis at Phytozome|http://www.phytozome.net/search.php?method=Org_Csinensis
The title that will apear in the sidebar appears before the ‘|’ charater. The link comes after. Now we want to add our downloads page. In the Resource Titles field add the text, ‘Downloads’, in the first field. Then in the first text area under Resource Blocks add the following text:
<p>The following annotation files are available for download:</p>
<b>Structural Annotations</b>
<table>
<tr>
<th>C. sinensis v1.0 scaffolds (FASTA format)</th>
<td><a href="/sites/default/files/citrus_sinensis-scaffold00001.fasta">citrus_sinensis-scaffold00001.fasta</a></td>
</tr>
<tr>
<th>C. siensis v1.0 genes sequences (FASTA format)</th>
<td><a href="/sites/default/files/citrus_sinensis-orange1.1g015632m.g.fasta">citrus_sinensis-orange1.1g015632m.g.fasta</a></td>
</tr>
<tr>
<th>C. siensis v1.0 genes (GFF3 format)</th>
<td><a href="/sites/default/files/citrus_sinensis-orange1.1g015632m.g.gff3">citrus_sinensis-orange1.1g015632m.g.gff3</a></td>
</tr>
</table>
<b>Functional Annotations</b>
<table>
<tr>
<th>Blast of C. sinensis v1.0 mRNA vs NCBI nr (XML format)</th>
<td><a href="/sites/default/files/blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_nr.out">blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_nr.out</a></td>
</tr>
<tr>
<th>Blast of C. sinensis v1.0 mRNA vs ExPASy SwissProt (XML format)</th>
<td><a href="/sites/default/files/blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_uniprot_sprot.fasta.out">blastx_citrus_sinensis-orange1.1g015632m.g.fasta.0_vs_uniprot_sprot.fasta.out</a></td>
</tr>
<tr>
<th>InterPro analysis of C. sinensis v1.0 mRNA (XML format)</th>
<td><a href="/sites/default/files/citrus_sinensis-orange1.1g015632m.g.iprscan.xml">citrus_sinensis-orange1.1g015632m.g.iprscan.xml</a></td>
</tr>
<tr>
<th>KEGG analysis of C. sinensis v1.0 mRNA (KEGG heir format)</th>
<td><a href="/sites/default/files/citrus_sinensis-orange1.1g015632m.g.KEGG.heir.tar.gz">citrus_sinensis-orange1.1g015632m.g.KEGG.heir.tar.gz</td>
</tr>
</table>
Click Save. Now, the link to Phytozome is present in the Resources sidebar and the Download block appears as follows:
Note:: Above we added HTML for our downloads block. However, if you prefer, the ckeditor module can be installed which can provide a Microsoft Word 2003 style interface which makes creation of content much easier without the need for entering HTML. HTML works well for this tutorial for quick cut-and-paste.
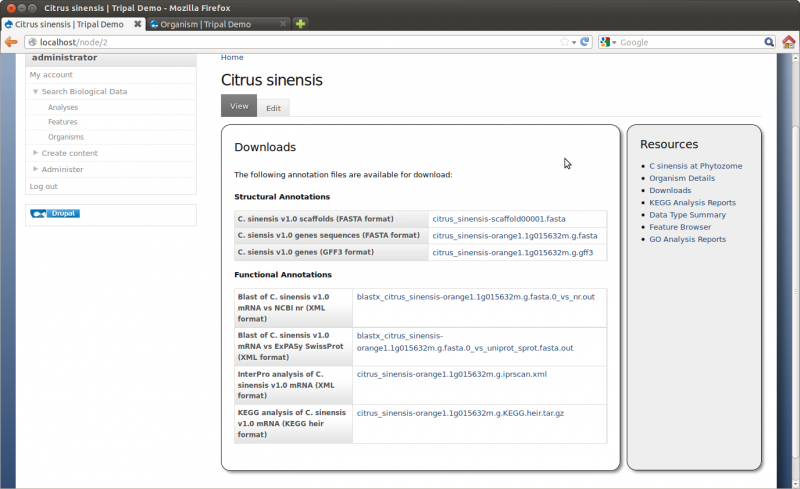
Because we set the fields to be unlimited in number you can add as many links or titles with blocks as you like by editing the organism page and adding more.
Finally, we can do the same for all Tripal content types (e.g. Analysis, Feature, etc.) However, because we have already created the CCK fields we don’t need to create them again. We simply reuse them on the other content types. For example, if we were to add these same fields to the Analysis content type, a new section appers when managing the fields called Existing field.
We can use this field to add all three fields we created previously to the Analysis content type. For consistency we should use the same name for the label.
Advanced Features
Custom and Search Pages (Views Integration)
Drupal Views is a powerful module that allows site administrators to create custom pages or search interfaces for data in the database tables accessible to Drupal. In Tripal v1.0 all Chado tables in both the Chado v1.1x and v1.2x schemas have been described to Drupal Views. This allows a site administrator to create custom pages for any type of Chado content regardless if it is supported by Tripal.
Tripal provides an interface called Views Integration which allows the site administrator to control how Chado tables are exposed to Views. A site administrator can use this interface to change the way data is handled in search forms and displayed on the page. This is an advanced feature and is not covered by this tutorial.
By default, Tripal comes with several default views. These are the search forms under the Search Biological Data link in the navigation menu. We have used these search forms several times in this tutorial to find an organism, feature or analysis. If you prefer different columns in the search results, or would like to expose different fields for search criteria, you can edit the View that corresonds to each of these search forms to change how these search pages behave. YOu edit the Views using the Views UI web interface and no PHP, HTML or any web programming is required to make changes to these search pages.
Additionally, you may use Views to create custom pages of Chado content. This is useful in the case that a specific data type is not supported by Tripal. Currently, there is no genetic marker page for Tripal. You could use views to query the database for marker features and displays information about them. Ideally, for complete control you would need to create a custom Drupal module to plug into Tripal that could display your data–especially if the visualization is complex. But using Materialized views, Views Integration and Drupal Views, you can create custom pages not currently supported by Tripal.
For infomation on working with Views see the Views project page: http://drupal.org/project/views. Rembmer that Tripal v1.0 runs on Drupal 6.x using Views 2 (no need to learn Views 3 on Drupal 7).
The Tripal Bulk Loader
Another advanced feature not described in this tutorial is the Tripal Bulk Loader, which is a new feature added to version 1.0. Often, data is not in common formats such as GFF, FASTA, GAF, InterPro XML, etc., but rather in Excel spreadsheets or tab-delimited or comma-separted files. The goal of the bulk loader is to enable a user to load data in these formats into the Chado schema. Currently, the bulk loader allows a site administrator to create custom loader templates that will allow a user to load tab-delimited files of any format.
Using the bulk loader web-interface, the priviledged user creates a “template” for loading a tab-delimited file. This templates specifies which fields in the Chado tables the values in the tab-delimited file will be stored. Once the template is fully defined, the priviledge user saves the template for other users to use. Another user can then load any tab-delimited files that matches the template. The user can upload as many files as desired.
A tutorial for using the Bulk Loader is under development and will be coming in the future.
Creating Custom Modules
As mentioned early in the Tutoral, Tripal is a modular software package. A Tripal API has been developed to help others who want to extend the functionality of Tripal. Anyone is welcome to develop modules for Tripal to suit their own needs and perhaps share them back with the community. The Tripal API can be found on the home page: http://tripal.sourceforge.net/. Information for developing new module can be found in the Tripal Developer’s Handbook.
Modules that conform the the Tripal API and Drupal coding standards will be officially approved by the Tripal Developers Consortium. These modules will be listed on the Tripal website and will be available in the Drupal module repository for download.
Anyone wishing to extend Tripal should sign up for the developers mailing list https://lists.sourceforge.net/lists/listinfo/gmod-tripal-devel and try to attend one of the monthly developer’s meetings to discuss the desired extensions.
Facts about “Tripal Tutorial (v1.0)”
| Has topic | Tripal + |