GMOD
Talk:Chado Natural Diversity Module Working Group
Contents
- 1 Responsibilities
- 2 January 2010
- 3 February 2010
- 4 March 2010
- 5 April 2010
- 6 simplified schema (ND tables only)
This is the discussion page for the Chado Natural Diversity Module Working Group. Notes on what we talk about and what decisions are made will be posted here. Eventually, as we settle on specific outcomes, those outcomes will be posted to the main page, and eventually reflected in the Chado schema.
At this time (January 2010), most of this discussion is about making changes relative to the version that was created in 2007 with Heliconius in mind. This is referred to below as HDB.
We will also use what came up at the PAG meeting as starting point for the discussion. This will change over time, as things settle down.
Responsibilities
Note: These responsibilities are flexible. They are just what we decided at the PAG meetings. They are wide open to discussion (or just add your name below.
Sook Jung will take the lead on schema changes/development. Sook is interested and is motivated to produce a working schema as soon as possible.
Dave Clements will lead documentation efforts. Dave will produce wiki documentation for the new tables. I hope to also create schema diagrams (probably using Power Architect). Dave is also keenly interested in how phenotypes are represented both in this module, and in the rest of the Chado.
Add your name here …
January 2010
Schema Change Logistics
We have conflicting goals here:
- Some people need this module yesterday.
- Some people want this module to strike the perfect balance between usability and flexibility.
Neither group is opposed to the other group’s goal per se - they just happened to be incompatible goals.
To address both these needs, Rob Buells proposed
- multiple incremental releases,
- with perhaps some backwards incompatability,
- but always with a plan and code support for migrating from one release to the next.
Rob points out that this might also be a good test case for establishing a formal process making backwards-incompatible updates to production Chado.
Schema Design Tools / Visualization
Sook, who is leading this particular effort, has access to Quest Toad for doing database design. She will likely send out proposals and revisions at Toad diagrams.
Dave, whose (who’s?) job is to document all this, can’t afford Toad. He will likely use Power*Architect to generate final as well as some intermediate documentation. Power*Architect is free, which is within my budget. (This also makes it possible for anyone else to update the doc in the long term.)
Observational Taxonomy
HDB has several different levels of biological unit, all represented with a different set of tables
- Organism - This already exists and comes from the Chado Organism Module. It defines a species.
- Biotype
- Stock (which is different from the stock table already in Chado)
- Individual
- Crossexperiment
- Specimen
And there are a bevy of relationships between these tables.
| Organism | M:M | Biotype | |
| Biotype | 1:M | Stock | there are 3 different 1:M rels |
| Stock | 1:M | Individual | |
| Crossexperiment | 1:M | Individual | |
| Individual | 1:M | Crossexperiemnt | |
| Individual | 1:M | Specimen | |
| Biotype | M:M | Individual |
All of this tables describe some unit/group of biology/life, ranging from species (organism) down to tissue in hand (specimen). The HDB design has several structurally identical tables in HDB for the various levels for different types of data (phenotype, images, …). This particular hierarchy is also particular to butterflies.
Stock
Both the HDB version and the production Chado have a stock table. The Chado Stock Module was added to production Chado while or after the HDB version was being developed.
The Chado Stock module is about keeping track of lines in your lab/community. Someone needs to take a look at it and determine how the natural diversity module should interact with it.
Observational Taxonomy Proposal
When Sook Jung mapped the HDB version to plant biology a number of issues came up:
- HDB associates genotypes with specimen_id, but in many cases genotypes are recorded at the level of individual trees, not part of the trees.
- The hierarchy of Biotype/Stock/Individual/Specimen requires storing of the same data multiple times (eg. all the organism entries need to be stored in biotype table since stock table has only stock.biotype_id).
- Multiple crosses can be done in one project (F1, BC1, and BC2 etc) and the individual cross experiment need to be linked to the larger project.
- ‘crossexperiment’ table may need to be linked to the project table by a linker table called ‘crossexperiment_project’, or
- We can have ‘project_relationship’ and ‘projectprop’ tables instead of ‘crossexperiment’ table to store data of various hierarchical projects (breeding experiments with multiple crosses and sub-trials, QTL projects with multiple crosses, etc).
This highlights that HDB is not a very Chadoesque design. We need to genericize the design to support arbitrary hierarchies, lineages, and mating types. This will support many more users and allow them to store images, phenotypes, genotypes, properties, etc. for whatever level of the hierarchy they have data for.
We can’t touch Organism, as it’s a key table in every Chado instance out there. However, everything else is open to change.
Observational Unit
The GDPDM has observational units, which represents whatever level of sample you have data for. I find that name descriptive, but awkward. Unfortunately, I can’t think of a better name. Suggestions are welcome.
Specifics:
- Try to combine biotype, stock, individual, and crossexperiemt into a single table, tentatively called obs_unit (with a nod to GDPDM).
- Investigate also folding specimen into obs_unit.
- An observational unit’s place in the observational taxonomy will be indicated by a new column in obs_unit that points to the CV table. For butterflies, the possible values might be “species”, “biotype”, “stock”, “individual”, and possibly “specimen”
Observational Unit Relationships
We need to support arbitrary M:M relationships between different levels of the observational taxonomy, and within the same level as well. For example, we may want a complete chain from species to individual (or plot or brood or …), and that individual may have resulted from crossing 2 other individuals (or from cloning one, or …).
The common solution is to create a bridge/mapping/intersection table to implement M:M relationships between obs_unit and itself. This table would define the standard “subject relationship object” triple where the subject and object are obs_unit’s and the relationship is a CV term.
This also deals with complications in lineage and mating types. You can represent T. Thermophila which has 7 mating types (any 2 will do), C. elegans which has hermaphrodites and outcrossing, E. coli which is asexual, …
It was pointed out that this table may contain cycles. In some experiments an individual will be crossed with it’s descendents. Therefore software that walks these relationships will need to detect cycles.
Current Status
We’re planning on moving ahead with merging the existing tables into a single table. And, no we can’t yet agree on that table name.
Project/Experiment/Study Hierarchy
The current Project table is defined in the General Module. The HDB design links to it extensively. However, other modules hardly use it at all.
The GDR group needs to the ability to more robustly define projects/studies, and to introduce substudies/project hierarchy, as well.
Phenotypes and Genotypes
Phenotypes
Phenotypes are not particularly well defined in Chado. Scott says that there are two sets of phenotype tables in Chado. One is a first rough draft that snuck in (and is used by some), and the other is a more robust set, which is used by others (including FlyBase). Too make things worse, which tables are in which set is not presently clearly defined.
Dave C. will do some research into
- What is currently going on in Chado?
- Which tables are in the old and new implementations?
- How are those tables currently used, and by whom?
- What are best practices for representing phenotypes in a generic database like Chado?
If items #2 and #1 don’t line up, and there are not a lot of current users, then I would like to look into
- reimplementing phenotypes in Chado, and
- providing migration paths for what users we do have.
HDB Design
The HDB version of the module ties into the preexisting phenotype table. The phenotype table has 4 foreign keys pointing to the cvterm table:
| observable_id | The entity: e.g. anatomy_part, biological_process. |
| attr_id | Phenotypic attribute (quality, property, attribute, character) - drawn from PATO. |
| cvalue_id | Phenotype attribute value (state). There is also a value text column which can hold unconstrained text. In any given record, exactly one of cvalue and value should have a non-null value. |
| assay_id | Evidence type |
Any of these 4 columns can be null.
There is also a phenotype_cvterm table to hold CV terms that don’t fit cleanly into the 4 CV term columns in phenotype.
It is not obvious to us (who aren’t that familiar with PATO), why the observable_id and attr_id columns are special enough to get their own permanent columns in the phenotype table. Shouldn’t they be thrown in phenotype_cvterm as well?
The current setup would work for:
Rohan-beard nerve cells show elevated response.
But would it work for
Rohan-beard nerve cells in the top of the dorsal fin show a faster and stronger response (Yes, I know this example may not make biological sense.)
Is this annotation fundamentally about Rohan-beard cells, the dorsal fin, the top of the dorsal fin, or all them put together? With the current design one of them has to come first.
As mentioned above, this area needs more research, or maybe just more documentation.
Genotypes
Genotypes appear to be more clearly defined in the production Chado schema: A genotype is a collection of features.
We discussed a particular use case: How would you store the results of an SSR assay? For example, in some individuals a given SSR is n bases long, while in another it is m bases long.
This would go in the genotype and feature tables. However, we may not be clear yet on where the length would go.
Functional and Sequence Alleles
Sook pointed out the need to represent both functional and sequence alleles. She explained: An apple geneticist who is working on a locus called F-M locus found three functional alleles and 12 sequence alleles.
- Functional: morphologically defined alleles (F, f and f1)
- Sequence: four SSR variants for F, six SSR variants for f, and two for f1 alleles.
We worked this out as functional alleles are stored in the phenotype tables, and sequence alleles in the feature and genotype tables.
Assays, Images
HDB includes support for images and assays. We should probably have a general purpose solution that is usable for all images and assays, not just those in the natural diversity module.
Action Items
Sook will
- write up some use cases
- generate a revised schema (perhaps just a draft), including a diagram
Dave will
- create a Doodle poll to determine the best time to meet. Poll is here.
- announce this group, this page, our next meeting, and the doodle poll to the GMOD Schema list.
Anything else?
February 2010
The first meeting in February will be held Monday February 8, at 11am Eastern US. Contact Dave C if you are interested in participating in this meeting.
Meetings after that will be scheduled at a regular time according to when people are usually available. Please fill out this Doodle poll to let us know when you can make it.
Scheduling this will be tough as we have key interested parties in Europe and across the contiguous US.
8 Feb 2010 Teleconference
March 2010
- New Proposal from Sook (PDF): Contains Use Cases and new/deleted/modified tables from HDB
- SQL
April 2010
Topic for Apr Teleconference Meeting
- The structure of diversityexperiment table and four different sub-tables
- Renaming tables
- ptexperiment/gtexperiment to ptassay/gtassay?
- ptassay/gtassay to ptprotocol/gtprotocol or ptassaytype/gtassaytype or ptmethod/gtmethod?
- diversityexperiment?
- Do we need crossexperiment table linked to stocksample table via diversityexperiment?
We (GDR) definitely need some sort of crossexperiment table linked to stock instead of stocksample
-
- crossexperiment (crossexperiment_id, name, expdate, experimenter_id, geolocation_id, type_id)
- crossexperiment_stock (crossexperiment_stock_id, crossexperiment_id, stock_id, type_id)
- crossexperimentprop (crossexperimentprop_id, crossexperiment_id, cvterm_id, value, rank)
- crossexperiment_project
If we need currrent crossexperiment table, we could name this new set of tables using cross- instead of crossexperiment-
- Adding diversityexperiment.collection_date
simplified schema (ND tables only)
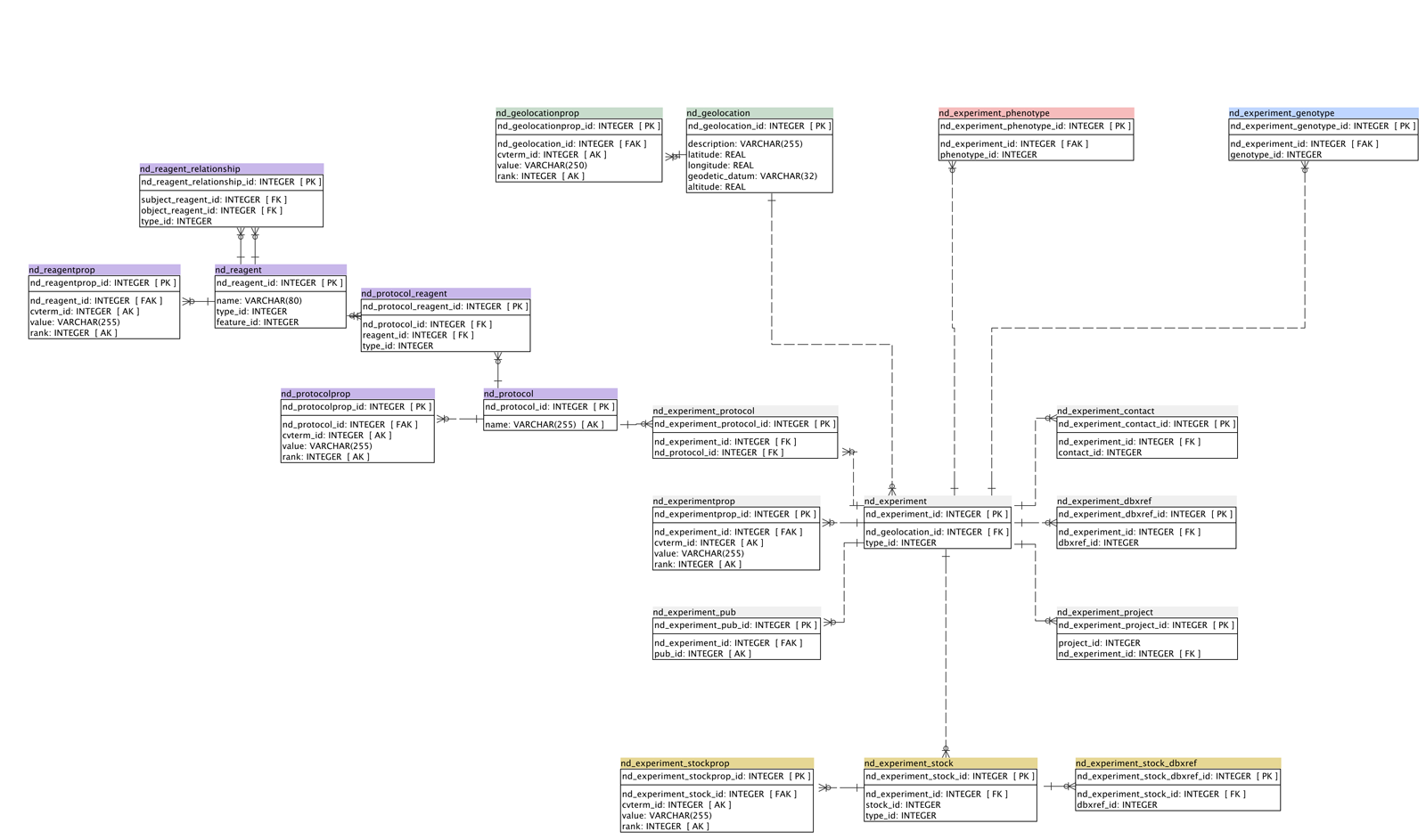
Notes on simplified nd schema and Use Cases