GMOD
PubFetch
The item described in this page is not (or is no longer) supported by the GMOD project.
This page is included only for reference purposes.
PubFetch is part of the PubSearch web-based literature curation toolset and functions as the interface between the literature curation tools and the online literature databases, such as PubMed. The aim of PubFetch is to provide a generic way of searching and retrieving literature data from online literature datasources so that the downstream applications dont have to deal with the idiosyncracies of the individual literature databases.
Initially PubFetch will act as the interface between PubSearch and the PubMed] and Agricola databases used by RGD and TAIR. A standard API and data format will be created to provide database queries and return results, popular existing formats and protocols will be used/supported wherever possible.
Documentation
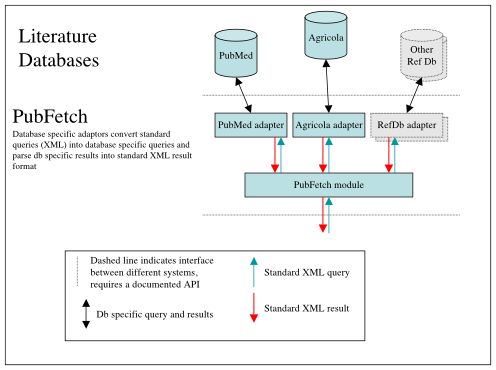
Figure 1 - Overview diagram of PubFetch showing how the PubFetch module will provide a generic literature access interface to PubMed and Agricola which could be expanded to other literature sources as desired.
The codebase will be developed initially in perl by adapting existing RGD perl modules designed to retrieve data from PubMed in a standard XML format. This code will be reviewed and adapted to create the main PubFetch module and appropriate database interace modules. Figure 2 below is a schematic diagram of the exising RGD literature download modules.
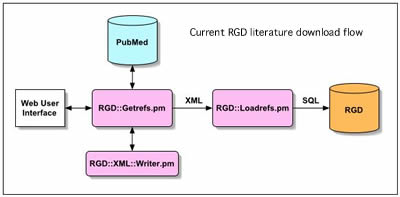
Figure 2- Current RGD literature download process showing perl modules used to interact with PubMed, create XML data and load into RGD.
The fundamental actions required of PubFetch are:
- Search LitDb for articles matching certain query criteria (eg.
keywords, date, author, etc).
- This will most likely entail passing the search critieria to PubFetch and retrieving a set of accession numbers (eg. PubMed IDs, PMIDs) for matching references.
- Retrieve the text information from the LitDb corresponding to a supplied accession number (e.g. bring me the PubMed entry for PMID 12345)
PubFetch as a BioMOBY webservice
To provide generic access to PubFetch we intend to make the core functionality available as a webservice, following the BioMOBY service model. The two actions described above will be implemented as two classes of Web services, the first taking keywords and returning PubMed IDs (or other LitDb accession) , the second taking LitDb accessions and returning the text information in a simple, standardized XML format. We will endeavour to provide the data in existing formats (raw data from the LitDb, a BioPerl-compatible format, etc) in addition to a simple XML format that is not dependent on other codebases
Downloads
The source code can be downloaded from SourceForge.