GMOD
PrimerDesigner.pm
PrimerDesigner is a GBrowse plugin, written by Sheldon McKay, that uses use the primer3 program to design PCR primers for selected regions within the genome browser. Please feel free to contact the author or The Gbrowse mailing list for help or more information.
The video below demonstrates designing PCR primers using the PrimerDesigner plugin.
Contents
- 1 How to use the PrimerDesigner plugin
- 2 How to install the PrimerDesigner plugin
- 3 PrimerDesigner and GBrowse 2
How to use the PrimerDesigner plugin
Accessing the plugin
- From GBrowse main page, the PrimerDesigner plugin, as well as other installed plugins, can be accessed via the ‘Reports and Analysis’ menu.
- In GBrowse, navigate to the genomic region you are interested in, then select ‘Design PCR primers’ from the menu and click ‘Configure’
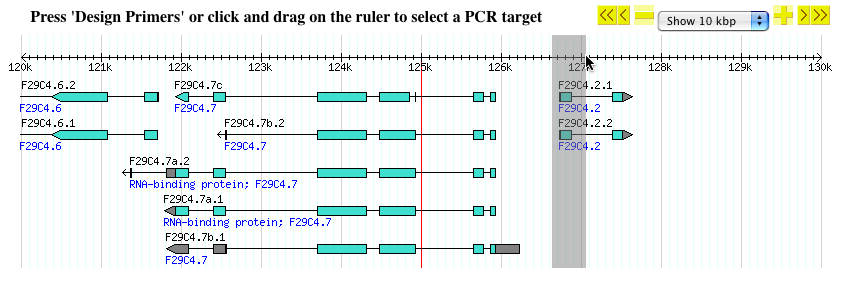
- You will then see a view similar to this:
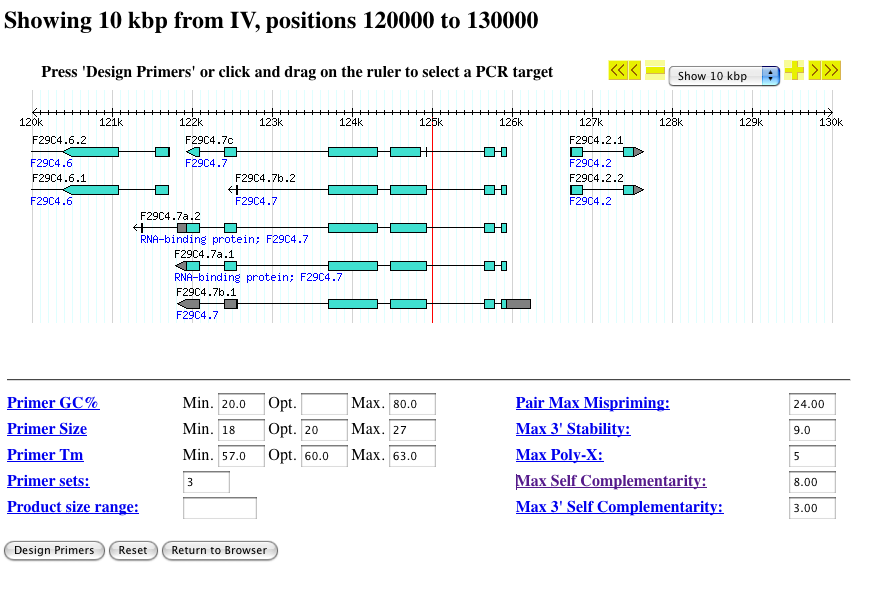
- At the top right corner is a navigation menu that allows you to zoom and pan the displayed segment in a manner similar to the main Gbrowse user interface.
Selecting the target for the PCR primers
- If no target region is specified, primers will be designed to amplify the center of the displayed sequence, indicated by a vertical red line.
- You can specify the target region for PCR primers using rubber-band selection.
- To activate this feature, point your mouse on the scale bar at the top of the image and click your mouse-button down, without releasing it.
- Then drag your mouse to the left or right to highlight the region or annotations you are interested in.
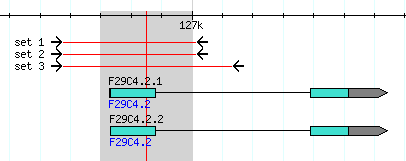
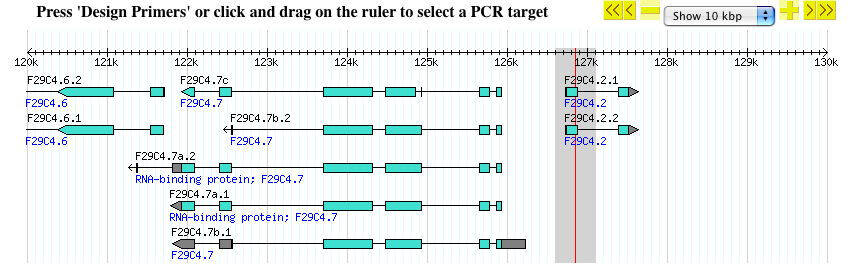
- In the example below, the first exon of gene F29C4.2 is highlighted with rubber-band selection.
- Once you are done selecting, release the mouse button and the page will reload with the selected region targeted for PCR primers
PCR primer design parameters
- Design parameters, such as primer length, GC content, etc can be configured at the bottom of the page.
- Each parameter can be clicked for detailed information.
- Reasonable default options are provided for each parameter. These options usually work but can be adjusted as desired.
- The size of the PCR product is calculated based on the size of the selected region. This can be manually adjusted, if desired.

Predicted PCR Primer Results
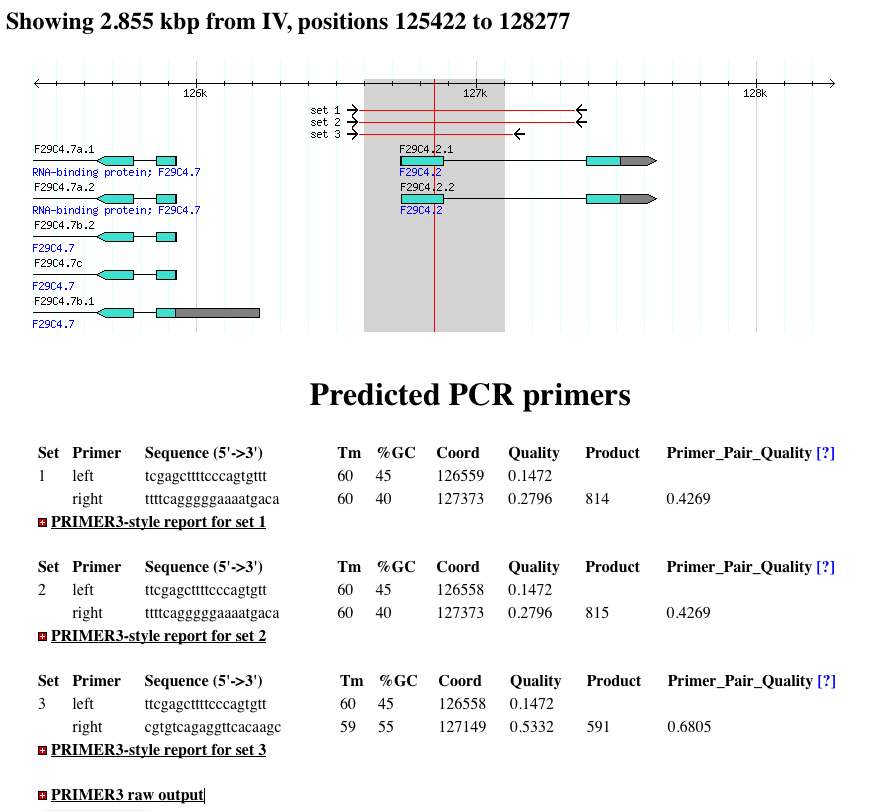
- Once you have clicked ‘Design Primers’, you will see a results page similar to the example below.
- The PCR primers are mapped back onto the image, so you can see the location of the predicted PCR amplicon.
- Below the image, primer statistics are show.
- You can also click to expand the sections marked with the [+] icon for more detailed reports.
Note On Quality Scores
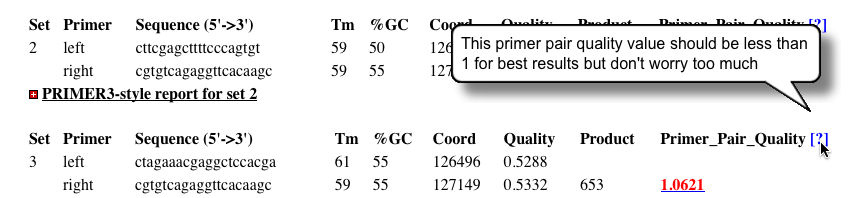
- Occasionally you will see quality scores flagged in red.
- Optimal primer scores should be less than 1
- Higher scores often result from minor departures from optimality criteria such as primer length, GC content, etc.
- If the primers do not fall to far outside of the requested parameters, it is not serious.
- However, being mindful that the primers are only as good as the available sequence will allow, making changes to the allowed product size range or target coordinates will permit more optimal primers to be selected.
How to install the PrimerDesigner plugin
First of all, you will need to install GBrowse, with all of its prerequisites, such as BioPerl and Bio::Graphics
Dependencies
Primer3
- Follow this link to go the primer website at Sourceforge.
- Primer3 is a binary executable, developed by Steve Rozen and colleagues, that the PrimerDesigner plugin uses to to the actual work of designing primers
- The plugin passes the DNA sequence and primer design parameters to this program and then parses the results
Installing Primer3
- Download or compile the suitable primer3 binary executable for your website.
- If necessary, change the name of the binary to ‘primer3’ so that it is recognized by the PrimerDesigner plugin
- The default path for primer is in ‘/usr/local/bin’. If you have installed it elsewhere, be sure to the the ‘binpath’ option (see below) in your configuration file.
Bio::PrimerDesigner
- Follow this link for information about the Bio::PrimerDesigner package.
- Bio::PrimerDesigner is a set of perl modules that provide a low-level API for the primer3 program.
- Note: Bio::PrimerDesigner is not part of BioPerl or GBrowse; you will have to install this package for the Primerdesigner plugin to work.
Installing Bio::PrimerDesigner
- Bio::PrimerDesigner is on CPAN (the Comprehensive Perl Archive Network).
- To install it, you can use the CPAN shell.
Configuration
The Configuration File
Activating the Plugin
To activate the Primer Designer plugin, simple add its name to the list of available plugins in the configuration file for you data source.
For example:
# Installed plugins
plugins = Spectrogram SequenceDumper PrimerDesigner
NOTE: For this plugin to work properly, you must ensure that all dependencies (described above) are that the GBrowse database has DNA sequence.
Configurable Options
To configure the plugin, add a stanza like this at the end of your [GENERAL] section:
[PrimerDesigner:plugin]
binpath = /usr/bin/
method = local
ispcr = http://genome.ucsc.edu/cgi-bin/hgPcr?db=cb3
binpath
The default value is ‘/usr/local/bin’
Use this option to specify where the primer3 binary is located, if not in the default ‘/usr/local/bin’
If you are using a remote URL (primer3 is installed on another server), specify a URL instead.
method
The default value is ‘local’
Recognized options are ‘local’ (primer3 installed on the same server as Gbrowse) or remote (primer3 is installed on another server).
The ‘remote’ method is implicit if the binpath option is set to a URL beginning with
‘http://’
default size
The default value is 10_000
This option specifies the default segment size to show in the PrimerDesigner image
max range
The default value is 300
This option species the degree to which the PCR product size can depart from the optimal size, as determined by the size of the selected target area for PCR primers.
ispcr
This option can be set to a URL that point to the UCSC in silico PCR
sight.
If selected, in silico PCR results will be reported in the output of this plugin
These results help to determine if the correct amplicon is found and also indicate whether other PCR products
are amplified by the primer pair.
You must check the site to see if there is a genome sequence for you species and modify the above example accordingly
PrimerDesigner and GBrowse 2
The PrimerDesigner plugin does not work with GBrowse 2.00. Work is currently underway to fix this before the next release of GBrowse.