GMOD
JBrowse Desktop
This page provides a reference guide for “Desktop JBrowse” or “JBrowse Desktop”, which is a electron application that was unveiled for the release of JBrowse 1.12.0.
The operation of JBrowse Desktop is comparable to other desktop genome browsers like IGV or IGB, and it can be run without setting up a webserver (most of the time normal jbrowse runs on apache and is viewed in a web browser). In contrast, JBrowse Desktop is a simple exe file that can easily be run locally on Windows, Mac OS, or Linux.
Contents
- 1 Installation
- 2 Overview of JBrowse Desktop
- 3 Screenshot
- 4 Limitations
- 5 Features
- 6 Technical details
- 7 Tested platforms
- 8 External Links
Installation
At the most basic level, setting up JBrowse Desktop consists of:
- Downloading a zip file script from jbrowse.org e.g. from http://jbrowse.org/jbrowse-1-12-1/ and OSX and a Windows version exsits JBrowse-1.12.1-desktop-osx.zip and JBrowse-1.12.1-desktop-win.zip
- After downloading unzip the file to somewhere. There is no official uninstaller but you can move it to the Program files or Applications directory as needed.
Note: on Mac, you can alternatively use Homebrew and run `brew cask install jbrowse` which will download and place jbrowse in your Application directory
Overview of JBrowse Desktop
When you run JBrowse Desktop you will see a sort of “welcome screen” that gives you several options
- View Volvox sample data
- Open sequence file, this allows you to open FASTA, Indexed FASTA, or .2bit
- Open data directory, this allows you to open a data directory that has been prepared with normal jbrowse tools
Note: it is highly recommended to use Indexed FASTA (selecting both a fasta and a fai file generated by `samtools faidx file.fasta`) or 2bit. Even small genomes with normal FASTA will be much slower!
Screenshot
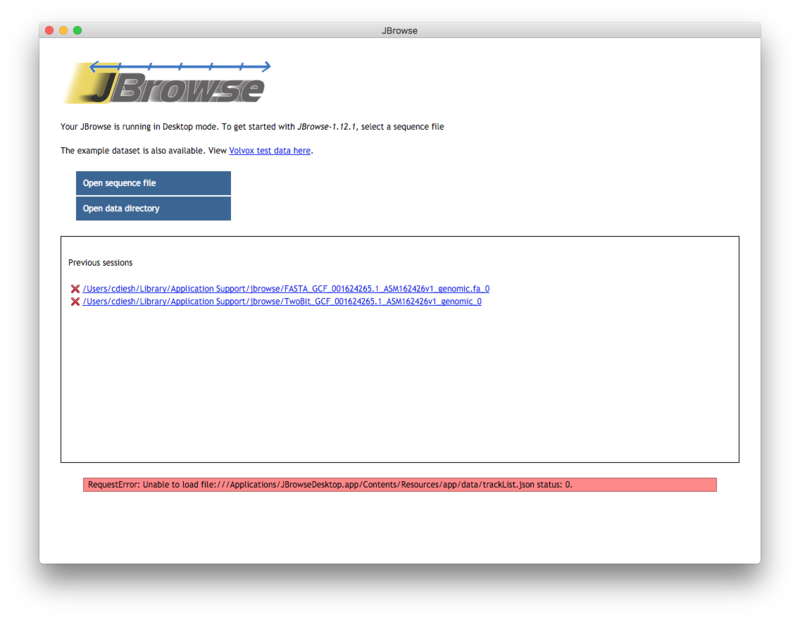
This is the welcome page when you open up the jbrowse app. You can open up sequence files such as fasta, 2bit, or indexed fasta here. Or, you can open up a pre-existing jbrowse data directory. You can also see a list of past “sessions” which can be revisited later. The red warning is sort of awkward but it does not have any meaning here, it just means that you aren’t currently in jbrowse main screen
Limitations
- The “Save session” function will not save some configuration customizations, so if you have advanced customizations it would be recommended to make a data directory by hand the trackList.json and tracks.conf format files
- If you open a data directory on the web, then “Save session” won’t work
Features
- Can open files from shared network folders and external hard drives on both windows and mac
- Can open files from remote URLs
- Can open data directories from remote URL
- Saves “sessions” to the OS “application data” folders
- Has a “home screen” that lists previous sessions
Technical details
The folder where sessions are saved is generally
- OSX - “~/Library/Application Support/jbrowse”
- Windows - “C:\Users\username\AppData\Roaming\jbrowse”
Tested platforms
- OSX Mavericks
- macOS sierra
- Windows 7
- Windows 10
Note: JBrowse Desktop doesn’t work on windows XP and Vista (all electron apps are limited to Windows 7+)