GMOD
Chado Natural Diversity Module
The Chado Natural Diversity Module is an extension to the Chado schema to better support natural diversity data.
Eventually this page will resemble the other Chado Module pages, with an overview followed by a detailed explanation of the tables, columns, and relationships in the module. However, while the module is under development, this page will have an alternative structure.
Recently, a lot of work was done on the ND module at the GMOD Tools for Evolutionary Biology hackathon, see https://www.nescent.org/wg_gmodevohackathon/Natural_Diversity_and_Phenotypes_Subgroup.
Contents
- 1 Introduction
- 2 Use Cases
- 3 ER Diagram
- 4
Tables
- 4.1 Table: nd_experiment
- 4.2 Table: nd_experiment_contact
- 4.3 Table: nd_experiment_dbxref
- 4.4 Table: nd_experiment_genotype
- 4.5 Table: nd_experiment_phenotype
- 4.6 Table: nd_experiment_project
- 4.7 Table: nd_experiment_protocol
- 4.8 Table: nd_experiment_pub
- 4.9 Table: nd_experiment_stock
- 4.10 Table: nd_experiment_stock_dbxref
- 4.11 Table: nd_experiment_stockprop
- 4.12 Table: nd_experimentprop
- 4.13 Table: nd_geolocation
- 4.14 Table: nd_geolocationprop
- 4.15 Table: nd_protocol
- 4.16 Table: nd_protocol_reagent
- 4.17 Table: nd_protocolprop
- 4.18 Table: nd_reagent
- 4.19 Table: nd_reagent_relationship
- 4.20 Table: nd_reagentprop
Introduction
The Natural Diversity module allows storing data from multiple experiments of the same accessions/lines/strains, collected, treated and evaluated in various locations, environments, and times. Each accessions/lines/strains can be scored for a large number of phenotypic traits, and genotyped with an array of genetic markers. In addition to storing data from experiments performed on existing accessions/lines/strains, the tables in this module and other interacting modules allow storing data from experiments that generate new accessions/lines/strains and experimental samples, such as field collection, cross, and treatments.
Interactions with Other Chado Modules
Stock Module
The original stock module was designed to store information about stock collections in a laboratory. This original concept of ‘stock’ table has been expanded to accommodate entities that a ‘stock’ belongs to or entities that derived from a ‘stock’. Hence ‘stock’ table can store hierarchical entity of population, strain/line/accession, individual, clone, and sample, with relationships between ‘stocks’ defined in the ‘stock_relationship’ table. For example, a plant accession can belong to a population (eg. progeny of a particular cross), and have multiple ‘children’ when used as parent in a cross. A plant accession can also have a ‘clone’ when it is clonally propagated. The ‘stock’ table also stores ‘samples’ or ‘observational units’ of individual experiments. For example, when a plant was treated with different amounts of chemicals before various phenotyping experiments were performed, each sample treated with specific amounts of chemicals is stored in a distinct row of ‘stock’ table. Since population can be defined as a group of any entities, a population entity can be composed of multiple species (eg. a group of insects collected in a field). To accommodate this change, the ‘NOT NULL’ constraint for organism_id has been dropped.
General Description of Natural Diversity Module
- This section can be removed later on from this wiki page since most of the content exists in the table descriptions below. But it is included here as a summary to help us write the Use Cases.
Nd_experiment is the core table for the natural diversity module, representing each individual assay that is undertaken (nb this is usually *not* an entire experiment). Experiment.type is a cvterm that will define which records are expected for other tables. The types of experiments that are done on or gave rise to ‘stocks’ include, but are not restricted to, cross, field collection, sample treatment, phenotyping and genotyping experiments. Any cvterm may be used but it was designed with terms such as: [phenotype_assay, genotype_assay, field_collection, cross_experiment, sample_treatment] in mind.
For experiments of type ‘phenotype_assay’ or ‘genotype_assay’, each nd_experiment generally give rise to a single genotype or phenotype. Though there is no longer a one-to-one restriction, it is expected that phenotypes and genotypes will be the result of a single assay. An exception to one-to-one relationship would include the genotype of heterozygotes where the genotype of each allele is stored in a distinct row of genotype table. One record of phenotype and genotype can be linked to multiple experiments since multiple stock entries can produce same phenotype and/or genotype.
Each nd_experiment of type ‘phenotype_assay’ or ‘genotype_assay’ should be linked to a single stock entry via nd_experiment_stock table. So when multiple samples from the same line/accession/strain have been used for multiple phenotype assays, a unique nd_experiment_id should be created for a unique combination of a sample and a phenotype assay. The sample treatment that are performed to generate a sample from a line/accession/strain can be stored in a separate row of nd_experiment of type ‘sample_treatment’. Collections of assays that relate to each other can be linked via the same record in the ‘project’ table and/or same record in ‘stock’ table.
For experiments of type ‘field_collection’ and ‘cross_experiment’, the appropriate stock entries that are used or generated from the experiment can be linked by nd_experiment_stock table. In cross experiment, the parental stocks and the progeny are stored in ‘stock’ table and their roles in cross can be recorded using an appropriate cvterm for nd_experiment_stock.type_id. The progeny of a cross and the stocks collected from a field collection can be stored in ‘stock’ table as a population or as an individual line/accession/strain/sample and then be linked to nd_experiment appropriately.
The detailed protocols of experiments can be described via one or more protocols. Reagents used in protocol can be stored in nd_reagent table. The same protocol and reagent can be linked to multiple experiments and protocols, respectively. Metadata of experiments, such as date and experimenters, can be stored as properties (nd_experimentprop).
Key Ontologies
Stock Relationship Ontology
Loading Data
There are, as yet, no standard flat file formats or loading scripts to load data into this module. Custom scripts will need to be written to insert your data in the database.
Web Front Ends
See Also
- Chado Natural Diversity Module Working Group page, and the group’s discussion page for background on how this module was created.
Email Threads
- Natural diversity module and phenotype cvterm values
- Use Cases on the simplified natural diversity module
- nd naming
Use Cases
tree fruit breeding data (tfGDR)
Cross Experiment
Data:
- Cross name/ID, location, female and male parent, progeny, project name, metadata such as how many seeds were produced, date of the cross, etc.
- Accession_ID, Aliases, cultivar name, pedigree, description
Chado:
- Accession_ID is stored in ‘stock’ table and the associated data such as cultivar name, pedigree, and description are stored in ‘stockprop’ table.
- nd_experiment.type_id is the cvterm_id for ‘cross_experiment’.
- Cross location is stored in ‘nd_experiment’ table (nd_geolocation_id).
- Cross name/ID and the all the metadata that are associated with the cross experiments are stored in ‘nd_experimentprop’ table (cvterm_id and value).
- Parent and progeny are stored in ‘stock’ table and they are linked to ‘nd_experiment’ via ‘nd_experiment_stock’ using type_id such as ‘is a female parent’, ‘is a progeny’, etc.
- The whole progeny is stored as a population in stock table and the individual line is linked to the population entry via ‘stock_relationship’ table.
- Individual crosses is linked to a larger project via nd_experiment_project.
Phenotype Assay
- Multiple clones of the same fruit tree accessions are planted in various lots of various orchards.
- The fruits are harvested from the tree in multiple times, freshly evaluated or stored in different conditions then evaluated for multiple phenotypes.
- The fruits of the multiple clonal trees in the same lot are combined for phenotype assays and are given the same clone_ID (?!!)
- Individual trees have a distinct repetition number (per individual tree) in addition to the clone_ID and the phenotypes of individual trees are evaulated multiple times.
Data:
- Sample_ID (given by tfGDR), Clone_ID (eg. wsu001_1, gala_1), Accession_ID (eg. wsu001, gala, etc), pick date, assay date, storage time, storage condition, evaluators, plant date, repetition number.
- rootstock_ID, site (orchard), sub_location (plot, row and position)
- Name, definition and scale of phenotype descriptor and phenotypic value
- An example of phenotype is ‘fruit size’ and their value can be 1 through 5 (1=very small; 2=small; 3=medium; 4=large; 5= very large)
Chado:
- Sample_ID is given by tfGDR for each sample to which a distinct set of phenotype assays were performed.
- Accession_ID, Clone_ID and Sample_ID are stored in ‘stock’ table and their relationship are stored in ‘stock_relationship’ table.
- A unique nd_experiment_id with type ‘phenotype_assay’ is created for a unique combination of a sample and its phenotype.
- pick date, assay date, storage time, storage condition, rootstock and any other sample properties are stored in stockprop table using cvterm and value.
- If more complicated treatments are performed on the sample (eg. fertilizers), nd_experiment of type ‘sample_treatment’ will be created.
- Evaluators of each phenotype experiment are stored in ‘contact’ table, and linked to ‘nd_experiment’ via ‘nd_experiment_contact’.
- Project information is stored in project, linked to nd_experiment via nd_experiment_project.
- Relationship between subprojects and larger projects are stored in project_relationship.
- Each breeder can have different definitions for similar phenotype descriptors, phenotype descriptors from each breeder are stored separately in ‘cvterm’ table (eg.fruit_size from a breeder called KE is stored as KE_fruit_size in cvterm table).
- The names and definitions of phenotype descriptors are stored in cvterm table and their scale is stored in ‘cvtermprop’ table using value and scale fields. For example, ‘KE_fruit_size’ has a scale of 1 to 5 with 1(tiny), 2(small), 3(medium), 4(large), and 5(very large), the numeric values are stored in cvtermprop.rank and the descriptions are stored in cvtermprop.value.
- Phenotype and the value , such as KE_fruit_size (attr_id) and 1 (value), are stored in phenotype table
Comments:
Tomato/potato breeder lines and cultivars (<a href=”http://solgenomics.net” class=”external text”
rel=”nofollow”>Sol genomics network</a>)</span>
Phenotyping experiment
A collection of 479 tomato accessions were used in several locations and years for assaying traits of breeding significance. The collection is an unstructured population of tomato varieties.
Chado Stock Module The stock table is used for storing any plant accession, collection, germplasm, or plants in a field plot. We use the stock_relationship table for defining those relationships. Each plant accession, and its derived germplasm, is stored in the stock table , with relationships between accessions defined in the stock_relationship table. For example, a plant accession can belong to a ‘parent’ population, and has multiple ‘children’, such as individual plants in a field plot.
In this data-set, we have in the stock table a Tomato cultivars population, and all the related accessions. The relationship between the accession stocks and the population stock is defined in stock_relationship (the population is the object in stock_relationship, and all accessions are subjects). Similar relationship is defined between each accession and the plants in the experiment’s field plots. For example accession Heinz 1706 belongs to the cultivars population, and has several field plots.
**Chado Natural Diversity Module** Each phenotyping event is stored in the nd_experiment table. An experiment, in the Natural Diversity module, is usually defined as a single genotyping event, or a phenotyping event performed on one or more plants, at the same time and location, for one or multiple traits. Then the experiment is linked with each participating stock (nd_experiment_stock table). In this example, the phenotyping was performed on plants in field plots, thus nd_experiment_stock links the experiment_id with the stock_id of the field plot. This allows storing multiple experiments of the same accessions, evaluated in different locations, environments, and years. Each plant can be scored for a large number of phenotypic traits, and genotyped with an array of genetic markers (see ‘SGN Genotyping experiment bellow).
Each phenotyping and genotyping experiment is also tied to a geolocation (nd_geolocation table). Metadata of experiments, such as date, environmental conditions, and person recording the data, are stored as properties (nd_experimentprop).
All the scored phenotype values and attributes are stored in the phenotype table, and genotypes in the genotype table. The 2 linking tables, nd_experiment_phenotype and nd_experiment_genotype, provide a 1-1 or 1-many relationship between phenotype/genotype and experiments.
Genotyping experiment
Each genotyping experiment (in this use case SNP and SSR markers were assayed, yielding a genotype of a SNP of number of repeats) has a unique nd_experiment_id, and is linked to the stock using nd_experiment_stock, and to the genotype using nd_experiment_genotype table.
ER Diagram
A simplified schema diagram by N. Menda and R. Buels
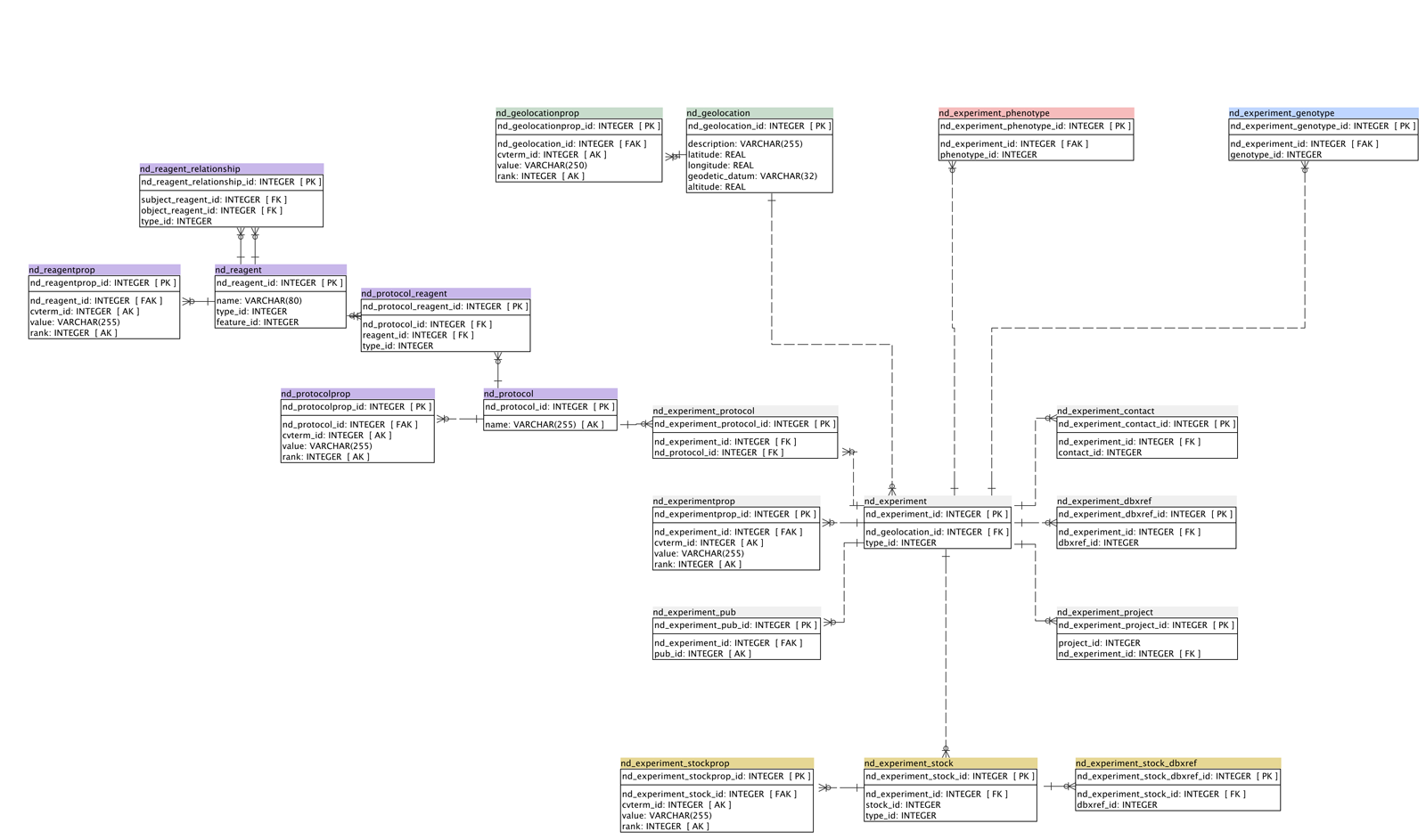
Tables
This will be populated using the process outlined in Chado Tables to Wiki.
Table: nd_experiment
This is the core table for the natural diversity module, representing
each individual assay that is undertaken (nb this is usually *not* an
entire experiment). Each nd_experiment should give rise to a single
genotype or phenotype and be described via 1 (or more) protocols.
Collections of assays that relate to each other should be linked to the
same record in the project table.
Experiment.type is a cvterm that will define which records are expected
for other tables. Any CV may be used but it was designed with terms such
as: [phenotype_assay, genotype_assay, field_collection,
cross_experiment] in mind.
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experiment_id | serial | PRIMARY KEY | |
| nd_geolocation | nd_geolocation_id | integer | NOT NULL |
| cvterm | type_id | integer | NOT NULL |
public.nd_experiment Structure
Tables referencing this one via Foreign Key Constraints:
Table: nd_experiment_contact
primary contact / submitter of these nd_experiments (nd, where assays are not submitted separately this may be better stored in project_contact).
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experiment_contact_id | serial | PRIMARY KEY | |
| nd_experiment | nd_experiment_id | integer | NOT NULL |
| contact | contact_id | integer | NOT NULL |
public.nd_experiment_contact Structure
Table: nd_experiment_dbxref
Cross-reference experiment to accessions, images, etc
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experiment_dbxref_id | serial | PRIMARY KEY | |
| nd_experiment | nd_experiment_id | integer | NOT NULL |
| dbxref | dbxref_id | integer | NOT NULL |
public.nd_experiment_dbxref Structure
Table: nd_experiment_genotype
Linking table: experiments to the genotypes they produce. Though there is no longer a one-to-one restriction, it is expected that genotypes will be the result of a single assay. The restriction was lifted to allow items such as: a single chromosome staining giving values for inversions: 2La/+, 2Rbc - (whilst these are technically a single genotype users may wish to store these separately such that all 2La/+ individuals can be easily ascertained).
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experiment_genotype_id | serial | PRIMARY KEY | |
| nd_experiment | nd_experiment_id | integer | UNIQUE#1 NOT NULL |
| genotype | genotype_id | integer | UNIQUE#1 NOT NULL |
public.nd_experiment_genotype Structure
Table: nd_experiment_phenotype
Linking table: experiments to the phenotypes they produce. in most cases this will either be a single record, or an alternative (quantitative / qualitative?) description of the same phenotype (e.g. 1: “wing length: 12mm” / “wing length: increased”). In rare cases it may suit the user to link a single qualitative phenotype to multiple experiments
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experiment_phenotype_id | serial | PRIMARY KEY | |
| nd_experiment | nd_experiment_id | integer | UNIQUE#1 NOT NULL |
| phenotype | phenotype_id | integer | UNIQUE#1 NOT NULL |
public.nd_experiment_phenotype Structure
Table: nd_experiment_project
Used to group together related nd_experiment records. All nd_experiments should be linked to at least one project.
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experiment_project_id | serial | PRIMARY KEY | |
| project | project_id | integer | NOT NULL |
| nd_experiment | nd_experiment_id | integer | NOT NULL |
public.nd_experiment_project Structure
Table: nd_experiment_protocol
Linking table: experiments to the protocols they involve.
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experiment_protocol_id | serial | PRIMARY KEY | |
| nd_experiment | nd_experiment_id | integer | NOT NULL |
| nd_protocol | nd_protocol_id | integer | NOT NULL |
public.nd_experiment_protocol Structure
Table: nd_experiment_pub
Linking nd_experiment(s) to publication(s)
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experiment_pub_id | serial | PRIMARY KEY | |
| nd_experiment | nd_experiment_id | integer | UNIQUE#1 NOT NULL |
| pub | pub_id | integer | UNIQUE#1 NOT NULL |
public.nd_experiment_pub Structure
Table: nd_experiment_stock
Part of a stock or a clone of a stock that is used in an experiment
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experiment_stock_id | serial | PRIMARY KEY | |
| nd_experiment_id | integer | NOT NULL | |
| stock_id | integer | NOT NULL stock used in the extraction or the corresponding stock for the clone |
|
| type_id | integer | NOT NULL |
public.nd_experiment_stock Structure
Tables referencing this one via Foreign Key Constraints:
Table: nd_experiment_stock_dbxref
Cross-reference experiment_stock to accessions, images, etc
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experiment_stock_dbxref_id | serial | PRIMARY KEY | |
| nd_experiment_stock | nd_experiment_stock_id | integer | NOT NULL |
| dbxref | dbxref_id | integer | NOT NULL |
public.nd_experiment_stock_dbxref Structure
Table: nd_experiment_stockprop
Property/value associations for experiment_stocks. This table can store the properties such as treatment
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experiment_stockprop_id | serial | PRIMARY KEY | |
| nd_experiment_stock_id | integer | UNIQUE#1 NOT NULL The experiment_stock to which the property applies. |
|
| type_id | integer | UNIQUE#1 NOT NULL The name of the property as a reference to a controlled vocabulary term. |
|
| value | character varying(255) | The value of the property. |
|
| rank | integer | UNIQUE#1 NOT NULL The rank of the property value, if the property has an array of values. |
public.nd_experiment_stockprop Structure
Table: nd_experimentprop
Tag-value properties - follows standard chado model.
| FK | Name | Type | Description |
|---|---|---|---|
| nd_experimentprop_id | serial | PRIMARY KEY | |
| nd_experiment | nd_experiment_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| value | character varying(255) | NOT NULL | |
| rank | integer | UNIQUE#1 NOT NULL |
public.nd_experimentprop Structure
Table: nd_geolocation
The geo-referencable location of the stock. NOTE: This entity is subject to change as a more general and possibly more OpenGIS-compliant geolocation module may be introduced into Chado.
| FK | Name | Type | Description |
|---|---|---|---|
| nd_geolocation_id | serial | PRIMARY KEY | |
| description | character varying(255) | A textual representation of the location, if this is the original georeference. Optional if the original georeference is available in lat/long coordinates. |
|
| latitude | real | The decimal latitude coordinate of the georeference, using positive and negative sign to indicate N and S, respectively. |
|
| longitude | real | The decimal longitude coordinate of the georeference, using positive and negative sign to indicate E and W, respectively. |
|
| geodetic_datum | character varying(32) | The geodetic system on which the geo-reference coordinates are based. For geo-references measured between 1984 and 2010, this will typically be WGS84. |
|
| altitude | real | The altitude (elevation) of the location in meters. If the altitude is only known as a range, this is the average, and altitude_dev will hold half of the width of the range. |
public.nd_geolocation Structure
Tables referencing this one via Foreign Key Constraints:
Table: nd_geolocationprop
Property/value associations for geolocations. This table can store the properties such as location and environment
| FK | Name | Type | Description |
|---|---|---|---|
| nd_geolocationprop_id | serial | PRIMARY KEY | |
| nd_geolocation_id | integer | UNIQUE#1 NOT NULL | |
| type_id | integer | UNIQUE#1 NOT NULL The name of the property as a reference to a controlled vocabulary term. |
|
| value | character varying(250) | The value of the property. |
|
| rank | integer | UNIQUE#1 NOT NULL The rank of the property value, if the property has an array of values. |
public.nd_geolocationprop Structure
Table: nd_protocol
A protocol can be anything that is done as part of the experiment.
| FK | Name | Type | Description |
|---|---|---|---|
| nd_protocol_id | serial | PRIMARY KEY | |
| name | character varying(255) | UNIQUE NOT NULL The protocol name. |
public.nd_protocol Structure
Tables referencing this one via Foreign Key Constraints:
Table: nd_protocol_reagent
| FK | Name | Type | Description |
|---|---|---|---|
| nd_protocol_reagent_id | serial | PRIMARY KEY | |
| nd_protocol | nd_protocol_id | integer | NOT NULL |
| nd_reagent | reagent_id | integer | NOT NULL |
| cvterm | type_id | integer | NOT NULL |
public.nd_protocol_reagent Structure
Table: nd_protocolprop
Property/value associations for protocol.
| FK | Name | Type | Description |
|---|---|---|---|
| nd_protocolprop_id | serial | PRIMARY KEY | |
| nd_protocol_id | integer | UNIQUE#1 NOT NULL The protocol to which the property applies. |
|
| type_id | integer | UNIQUE#1 NOT NULL The name of the property as a reference to a controlled vocabulary term. |
|
| value | character varying(255) | The value of the property. |
|
| rank | integer | UNIQUE#1 NOT NULL The rank of the property value, if the property has an array of values. |
public.nd_protocolprop Structure
Table: nd_reagent
A reagent such as a primer, an enzyme, an adapter oligo, a linker oligo. Reagents are used in genotyping experiments, or in any other kind of experiment.
| FK | Name | Type | Description |
|---|---|---|---|
| nd_reagent_id | serial | PRIMARY KEY | |
| name | character varying(80) | NOT NULL The name of the reagent. The name should be unique for a given type. |
|
| type_id | integer | NOT NULL The type of the reagent, for example linker oligomer, or forward primer. |
|
| feature_id | integer | If the reagent is a primer, the feature that it corresponds to. More generally, the corresponding feature for any reagent that has a sequence that maps to another sequence. |
public.nd_reagent Structure
Tables referencing this one via Foreign Key Constraints:
Table: nd_reagent_relationship
Relationships between reagents. Some reagents form a group. i.e., they are used all together or not at all. Examples are adapter/linker/enzyme experiment reagents.
| FK | Name | Type | Description |
|---|---|---|---|
| nd_reagent_relationship_id | serial | PRIMARY KEY | |
| subject_reagent_id | integer | NOT NULL The subject reagent in the relationship. In parent/child terminology, the subject is the child. For example, in "linkerA 3prime-overhang-linker enzymeA" linkerA is the subject, 3prime-overhand-linker is the type, and enzymeA is the object. |
|
| object_reagent_id | integer | NOT NULL The object reagent in the relationship. In parent/child terminology, the object is the parent. For example, in "linkerA 3prime-overhang-linker enzymeA" linkerA is the subject, 3prime-overhand-linker is the type, and enzymeA is the object. |
|
| type_id | integer | NOT NULL The type (or predicate) of the relationship. For example, in "linkerA 3prime-overhang-linker enzymeA" linkerA is the subject, 3prime-overhand-linker is the type, and enzymeA is the object. |
public.nd_reagent_relationship Structure
Table: nd_reagentprop
| FK | Name | Type | Description |
|---|---|---|---|
| nd_reagentprop_id | serial | PRIMARY KEY | |
| nd_reagent | nd_reagent_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| value | character varying(255) | ||
| rank | integer | UNIQUE#1 NOT NULL |
public.nd_reagentprop Structure