GMOD
CMAE
The item described in this page is not (or is no longer) supported by the GMOD project.
This page is included only for reference purposes.
Work on CMAE was started, but never completed.
The CMap Assembly Editor (CMAE) is a desktop application that is being developed to assist in visualizing and editing large scale sequence assemblies for the maize sequencing project. Using the CMap comparative mapping database, CMAE allows sequence assemblies to be superimposed on top of diverse other types of mapping data, allowing the finisher to view assemblies in the context of a cascade of mapping data at a variety of resolutions. For example, the editor can show sequence contigs aligned to fingerprinted physical map contigs, which are aligned in turn to genetic maps. Correspondence links between the different objects indicate the quality of the assembly and highlight possible mis-assemblies. The editor will then allow mis-assembled contigs to be split, merged or moved, or the troubled contigs can be exported to a more specialized program. (L. Stein)
Contents
Requirements
Documentation
Screenshots
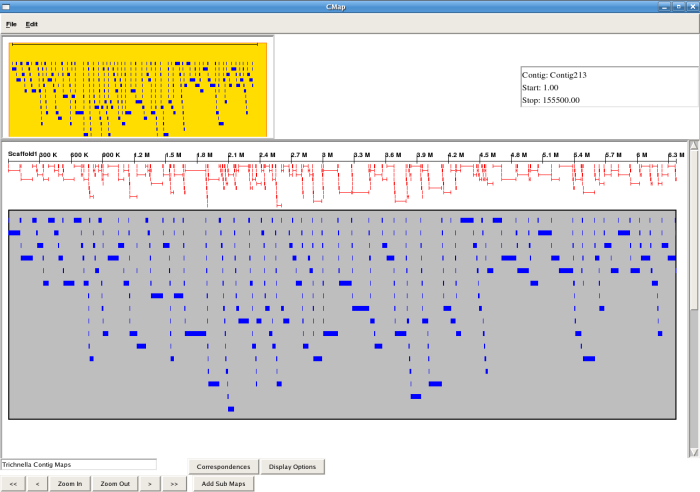
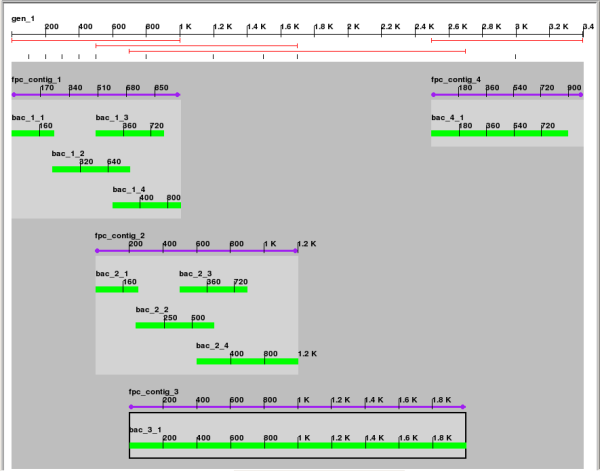
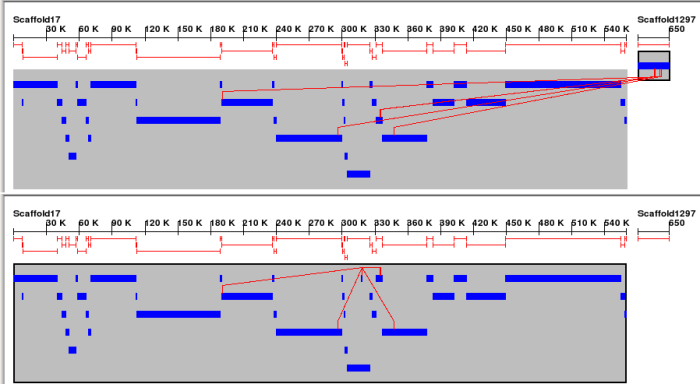
Code
The CMAE code is located in the SourceForge CVS repository for CMap.
The CMAE specific information is located in the “editor” directory.
Mailing List
- gmod-cmae@lists.sourceforge.net: Discussion of development, installation problems, etc.