Difference between revisions of "GBrowse syn"
From GMOD
(→Notes on Whole Genome Alignment Data) |
|||
| Line 7: | Line 7: | ||
=Notes on Whole Genome Alignment Data= | =Notes on Whole Genome Alignment Data= | ||
| − | + | The focus of this documentation is on the GBrowse_syn application. However, how to generate whole genome alignments, identify orthologous regions, etc, are the subject of considerable interest, so some background reading is listed below: | |
| − | The focus of | + | |
*<span class="pops">[http://www.eecs.berkeley.edu/Pubs/TechRpts/2006/EECS-2006-104.html Primer on Hierarchical Genome Alignment Strategies]</span> | *<span class="pops">[http://www.eecs.berkeley.edu/Pubs/TechRpts/2006/EECS-2006-104.html Primer on Hierarchical Genome Alignment Strategies]</span> | ||
*<span class="pops">[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2577869 article on PECAN and ENREDO]</span> | *<span class="pops">[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2577869 article on PECAN and ENREDO]</span> | ||
| Line 15: | Line 14: | ||
* The alignment data are in a <span class="pops">[http://gmod.org/wiki/GBrowse_syn_Database#Clustal_alignment_format constrained CLUSTALW format]</span> (They were not generated by the program CLUSTALW, whihc is not suitable for whole genome alignments) | * The alignment data are in a <span class="pops">[http://gmod.org/wiki/GBrowse_syn_Database#Clustal_alignment_format constrained CLUSTALW format]</span> (They were not generated by the program CLUSTALW, whihc is not suitable for whole genome alignments) | ||
* There are processing steps for the alignment data but it is very computationally intensive and we will load pre-processed data to get a head start. | * There are processing steps for the alignment data but it is very computationally intensive and we will load pre-processed data to get a head start. | ||
| − | |||
=Documentation= | =Documentation= | ||
Revision as of 09:58, 5 August 2009
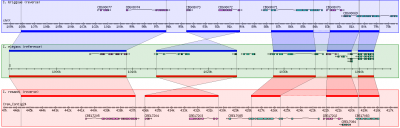
GBrowse_syn, or the Generic Synteny Browser, is a GBrowse-based synteny browser designed to display multiple genomes, with a central reference species compared to two or more additional species. It can be used to view multiple sequence alignment data, synteny or co-linearity data from other sources against genome annotations provided by GBrowse. GBrowse_syn is included with the standard GBrowse package (version 1.69 and later). Working examples can be seen at TAIR, WormBase, and SGN.
Contents
Notes on Whole Genome Alignment Data
The focus of this documentation is on the GBrowse_syn application. However, how to generate whole genome alignments, identify orthologous regions, etc, are the subject of considerable interest, so some background reading is listed below:
- Primer on Hierarchical Genome Alignment Strategies
- article on PECAN and ENREDO
- all about PECAN
- The gene annotations for each species are in GFF files.
- The alignment data are in a constrained CLUSTALW format (They were not generated by the program CLUSTALW, whihc is not suitable for whole genome alignments)
- There are processing steps for the alignment data but it is very computationally intensive and we will load pre-processed data to get a head start.
Documentation
Installation
- GBrowse_syn uses much of the same infrastructure as GBrowse, in terms of species databases, configuration files, perl libraries, etc.
- It comes as part of the GBrowse distribution, version 1.69 and later (GBrowse installation info...).
- It differs in that databases and configuration for individual species are linked together via a central configuration file and a joining database that contains reciprocal alignments between all species represented in the browser.
- We recommend using the most up-to-date version of the application either with the GBrowse netinstall script (use the -d option) or with a source code installation.
Configuration
- Configuration of GBrowse_syn is much the same as for GBrowse, with database and display options controlled by a configuration file
- GBrowse_syn uses a main configuration file for general options plus an individual configuration for each species represented in the mutliple sequence alignments
- Details of GBrowse_syn configuration can be found on this page.
The Alignment data
- GBrowse_syn uses a central 'joining' database that contains information about the multiple sequence alignments
- There is an additional database for each species represented in the alignments
- The species' databases are configured in the same way as a regular GBrowse installation
- Detailed information about the alignment database can be found on this page
User interface
- GBrowse_syn uses a central "reference species" panel, with inset panels above and below for two or more aligned species
- Details of the user interface can be found on this page
Presentations and Workshops
- Comparative Genomics with GBrowse_syn - Presentation by Sheldon McKay at the SMBE 2009 GMOD Workshop on using GBrowse_syn for comparative genomics.
- GBrowse_syn PAG 2009 Workshop
- November 2007 - Sheldon McKay's presentation on GBrowse_syn at the November 2007 GMOD Meeting.