|
|
| (9 intermediate revisions by 2 users not shown) |
| Line 1: |
Line 1: |
| − | {{SessionHead}}
| |
| − | {| class="tutorialheader"
| |
| − | | {{TutorialTitleLine|[[gmod:GBrowse_syn|GBrowse_syn]]}}<br />
| |
| − | [[2011 GMOD Spring Training]]<br />
| |
| − | 8-12 March 2011<br />
| |
| − | [[User:Mckays|Sheldon McKay, iPlant Collaborative, University of Arizona]]
| |
| − | | align="right" | {{#icon: GBrowse_synLogo.png|GBrowse_syn|200|gmod:GBrowse_syn}}
| |
| − | |}
| |
| | | | |
| | | | |
| | + | [[File:GBrowse_syn_logo.png|center|400px|alt=GBrowse_syn logo]] |
| | | | |
| − | <div style="float:right"> | + | <div class="componentBox"> |
| − | <center>[[Image:Gbrowse_syn.png|thumb|300px|GBrowse_syn as it looks at The Arabidopsis Information Resource (TAIR)]]</center> | + | <div class="compBoxHdr">Status</div> |
| − | {{SessionHead}} | + | *Mature release |
| | + | *maintenance |
| | + | *active support |
| | + | <div class="compBoxHdr">Resources</div> |
| | + | *[http://sourceforge.net/projects/gmod/files/Generic%20Genome%20Browser/ Download] |
| | + | *{{2008SurveyLink|#GBrowse_syn|2008 Survey}} |
| | + | *[[GBrowse_syn Tutorial]] |
| | </div> | | </div> |
| | | | |
| − | [[gmod:GBrowse_syn|GBrowse_syn]] is a [[gmod:GBrowse|GBrowse]]-based [[gmod:Synteny|synteny]] browser designed to display multiple genomes, with a central reference species compared to two or more additional species. It is included with the standard GBrowse package (version 1.69 and later).
| + | ==About Generic Synteny Browser (GBrowse_syn)== |
| − | *Working examples of GBrowse_syn can be seen at <span class="pops">[http://www.arabidopsis.org/cgi-bin/gbrowse_syn/arabidopsis/?name=Chr1%3A8367000..8370501;search_src=thaliana TAIR]</span> and <span class="pops">[http://dev.wormbase.org/db/seq/gbrowse_syn/compara?search_src=Cele;name=X:1050001..1150000 WormBase]</span>.
| + | |
| | | | |
| | + | GBrowse_syn, or the Generic Synteny Browser, is a [[GBrowse]]-based [[synteny]] browser designed to display multiple genomes, with a central reference species compared to two or more additional species. It can be used to view multiple sequence alignment data, synteny or co-linearity data from other sources against genome annotations provided by GBrowse. GBrowse_syn is included with the standard GBrowse package (version 1.69 and later). |
| | | | |
| − | __TOC__
| |
| | | | |
| − | =GBrowse_syn Introduction= | + | ===Screenshots=== |
| − | * An introductory talk will be presented using the slides below. Click the section to open.
| + | |
| | | | |
| − | <div class=switch id="Introductory Slides">
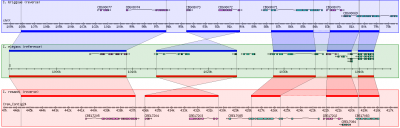
| + | [[Image:GBrowse_syn.png|none|400px|GBrowse_syn, as implemented at WormBase]] |
| − | [[Image:slide1.png|border]]<br><br> | + | |
| − | [[Image:slide2.png|border]]<br><br>
| + | |
| − | [[Image:slide3.png|border]]<br><br>
| + | |
| − | [[Image:slide4.png|border]]<br><br>
| + | |
| − | [[Image:slide5.png|border]]<br><br>
| + | |
| − | [[Image:slide6.png|border]]<br><br>
| + | |
| − | [[Image:slide7.png|border]]<br><br>
| + | |
| − | [[Image:slide8.png|border]]<br><br>
| + | |
| − | [[Image:slide9.png|border]]<br><br>
| + | |
| − | [[Image:slide10.png|border]]<br><br>
| + | |
| − | [[Image:slide11.png|border]]<br><br>
| + | |
| − | [[Image:slide12.png|border]]<br><br>
| + | |
| − | [[Image:slide13.png|border]]<br><br>
| + | |
| − | [[Image:slide14.png|border]]<br><br>
| + | |
| − | [[Image:slide15.png|border]]<br><br>
| + | |
| − | [[Image:slide16.png|border]]<br><br>
| + | |
| − | [[Image:slide17.png|border]]<br><br>
| + | |
| − | [[Image:slide18.png|border]]<br><br>
| + | |
| − | [[Image:slide19.png|border]]<br><br>
| + | |
| − | [[Image:slide20.png|border]]<br><br>
| + | |
| − | [[Image:slide21.png|border]]<br><br>
| + | |
| − | [[Image:slide22.png|border]]<br><br>
| + | |
| − | [[Image:slide23.png|border]]<br><br>
| + | |
| − | [[Image:slide24.png|border]]<br>
| + | |
| | | | |
| − | </div>
| + | ==Downloads== |
| | | | |
| | + | Download GBrowse_syn: [http://sourceforge.net/projects/gmod/files/Generic%20Genome%20Browser/ http://sourceforge.net/projects/gmod/files/Generic%20Genome%20Browser/] |
| | | | |
| − | =Installing GBrowse_syn=
| |
| − | GBrowse_syn is part of the GBrowse 2.0 package and was pre-installed when you went through the [[gmod:GBrowse 2.0 HOWTO|GBrowse 2.0 installation]].
| |
| | | | |
| − | <div class="emphasisbox" style="width:700px;margin-top:50px">
| + | The development version of GBrowse_syn is found at https://github.com/GMOD/GBrowse. Please be aware that development versions may have new features that are not fully tested. |
| − | <font color=red><b>Update:</b></font>
| + | |
| − | We will need to update the GBrowse source to include features and bug patches not included in the CPAN distribution:
| + | |
| − | </div>
| + | |
| − | * First, update {{CPAN|Bio::Graphics}}
| + | |
| − | <div class="indent">
| + | |
| − | $ <span class="enter">sudo cpan -i Bio::Graphics</span>
| + | |
| − | </div>
| + | |
| − | * Then check out a fresh copy of the current GBrowse 2 source code via [[gmod:SVN|subversion (svn)]].
| + | |
| − | <div class="indent">
| + | |
| − | $ <span class="enter">cd /home/gmod/Downloads/sources</span>
| + | |
| − | $ <span class="enter plainlinks">svn co https://gmod.svn.sourceforge.net/svnroot/gmod/Generic-Genome-Browser/trunk Generic-Genome-Browser</span>
| + | |
| − | $ <span class="enter">cd Generic-Genome-Browser</span>
| + | |
| − | $ <span class="enter">sudo perl Build.PL</span>
| + | |
| − | $ <span class="enter">sudo ./Build install</span>
| + | |
| | | | |
| − | NOTE: use the default options when prompted.
| + | ==Using GBrowse_syn== |
| − | </div>
| + | |
| − | Now point your browser to http://localhost/cgi-bin/gb2/gbrowse_syn
| + | |
| | | | |
| − | [[Image:welcome.png|border|thumb|left|600px|This is the welcome screen you should see after installing a new copy of GBrowse_syn with no configured data sources. It contains instructions on how to set up the example data source provided with the distribution.]] | + | GBrowse_syn has been part of the [[GBrowse]] distribution since version 1.69; we recommend using the most up-to-date version of GBrowse 2. Please follow the [[GBrowse_2.0_Install_HOWTO|installation instructions for GBrowse]]. |
| − | <br clear=all>
| + | |
| | | | |
| − | =Setting up the sample data= | + | ===Documentation=== |
| − | <div class="emphasisbox" style="width:700px;margin-top:50px">
| + | |
| − | * Sample data and configuration information for GBrowse_syn come pre-packaged with GBrowse.
| + | |
| − | * The example we will use is a two-species comparison of rice (''Oryza sativa'') and one of its wild relatives*
| + | |
| | | | |
| − | <font style="font-size:7pt">''*Data courtesy of Bonnie Hurwitz; sequences and names have been obfuscated to protect unpublished data''</font>
| + | See the [[GBrowse_syn_Help|help for GBrowse_syn]] |
| − | </div>
| + | |
| | | | |
| − | ==Setting up the Alignment Database== | + | ====Alignment data==== |
| | | | |
| − | The alignment, or joining database will contain the sequence alignments between the two rice species. It will be in a [[gmod:MySQL|MySQL]] database.
| + | * GBrowse_syn uses a central 'joining' database that contains information about the multiple sequence alignments |
| | + | * There is an additional GBrowse database for each species represented in the alignments |
| | + | * The databases for each species are configured in the same way as a regular GBrowse installations |
| | + | * [[GBrowse_syn_Database|Details on the GBrowse_syn database]] |
| | | | |
| − | 1) Create a MySQL database to hold the alignment data
| + | ====User interface==== |
| − | <div class="indent">
| + | |
| − | $ <span class="enter">mysql -u root -p</span>
| + | |
| − | Enter password: <span class="enter">****************</span>
| + | |
| − | Welcome to the MySQL monitor. Commands end with ; or \g.
| + | |
| − | Your MySQL connection id is 37
| + | |
| − | Server version: 5.1.37-1ubuntu5.1 (Ubuntu)
| + | |
| − |
| + | |
| − | Type 'help;' or '\h' for help. Type '\c' to clear the current input statement.
| + | |
| − |
| + | |
| − | mysql> <span class="enter">create database rice_synteny;</span>
| + | |
| − | Query OK, 1 row affected (0.00 sec)
| + | |
| − |
| + | |
| − | mysql>
| + | |
| − | </div>
| + | |
| | | | |
| − | 2) Give read-only (SELECT privileges in SQL) to the default apache user <tt>www-data</tt>. We can do this for all of the MySQL databases, since they are all for web applications
| + | The overall look of Gbrowse_syn resembles GBrowse but has some key differences to accomodate the more complex comparative genome data (see the [[Screenshot|screenshot]] above). |
| − | <div class="indent">
| + | |
| − | mysql> <span class="enter">GRANT SELECT on *.* TO 'www-data'@'localhost';</span>
| + | |
| − | Query OK, 0 rows affected (0.00 sec)
| + | |
| − |
| + | |
| − | mysql> <span class="enter">quit</span>
| + | |
| − | </div>
| + | |
| | | | |
| − | 3) Decompress the sample alignment data and load the database. You need to have root-level access (be a sudoer) for some of the steps below.
| + | GBrowse_syn uses a central "reference species" panel, with inset panels above and below for two or more aligned species. There is no upper limit to the number of species that can be displayed. |
| − | <div class="indent">
| + | |
| − | $ <span class="enter">cd /var/www/gbrowse2/databases/gbrowse_syn/alignments/</span>
| + | |
| − | $ <span class="enter">sudo gunzip rice.aln.gz</span>
| + | |
| − | Have a look at the first few lines of the data:
| + | |
| − | $ <span class="enter">head -20 rice.aln</span>
| + | |
| − | CLUSTAL W(1.81) multiple sequence alignment W(1.81)
| + | |
| − |
| + | |
| − |
| + | |
| − | rice-3(+)/16598648-16600199 ggaggccggccgtctgccatgcgtgagccagacggggcgggccggagacaggccacgtgg
| + | |
| − | wild_rice-3(+)/14467855-14469373 gggggccgg------------------------------------agacaggccacgtgg
| + | |
| − | ** ****** ***************
| + | |
| − |
| + | |
| − |
| + | |
| − | rice-3(+)/16598648-16600199 ccctgccccgggctgttgacccactggcacccctgtcccgggttgtcgccctcctttccc
| + | |
| − | wild_rice-3(+)/14467855-14469373 ccctgccccgggctgttgacccactggcacccctgtcccgggttgtcgccctcctttccc
| + | |
| − | ************************************************************
| + | |
| − |
| + | |
| − |
| + | |
| − | rice-3(+)/16598648-16600199 cgccatgctctaagtttgctcctcttctcgaacttctctctttgattcttcacgtcctct
| + | |
| − | wild_rice-3(+)/14467855-14469373 cgccatgctctaagtttgctcctcttctcgaacttctctctttgattcttcacgtcctct
| + | |
| − | ************************************************************
| + | |
| − |
| + | |
| − |
| + | |
| − | rice-3(+)/16598648-16600199 tggagcctccccttctagctcgatcacgctctgctcttccgcttggaggctggcaaaact
| + | |
| − | wild_rice-3(+)/14467855-14469373 tggagcctccccttctagctcgatcgcgctctgctcttccgcttggaggctggcaaaact
| + | |
| − |
| + | |
| − | The format is CLUSTALW. This is a formatting convention; it does not mean CLUSTALW was used to generate the alignment data. See [[#Further Reading|Further Reading]] below for more information on data loading and the meta-data in the sequence names
| + | |
| − | </div>
| + | |
| | | | |
| − | 4) Load the database with the script <tt>gbrowse_syn_load_alignments_msa.pl</tt>, which is automatically installed along with GBrowse. See the <span class=pops>[http://gmod.org/wiki/GBrowse_syn_Scripts GBrowse_syn scripts]</span> page for details on the options for the script.
| + | ==Publications, Tutorials, and Presentations== |
| − | <div class="indent">
| + | |
| − | $ <span class="enter">gbrowse_syn_load_alignments_msa.pl -u root -p gmodamericas2011 -d rice_synteny -c -v rice.aln</span>
| + | |
| | | | |
| − | There are 1800 alignment blocks, so this will take a little while to run.
| + | ===Publications on or mentioning GBrowse_syn=== |
| − | </div>
| + | |
| | | | |
| − | ==Setting up the Configuration Files==
| + | Please refer to the following paper when citing GBrowse_syn: |
| − | <div class="emphasisbox">
| + | |
| − | * The configuration files required for this data source are pre-installed with [[GBrowse]], in <tt>/etc/gbrowse2/synteny/</tt>.
| + | |
| − | * There are two species' config files, <tt>rice_synteny.conf</tt> and <tt>wild_rice_synteny.conf</tt>, and the joining config file, <tt>oryza.synconf</tt>. The latter file has been disabled by appending a '.disabled' extension to the file name.
| + | |
| − | </div>
| + | |
| − | The joining config file, <tt>oryza.synconf</tt>:
| + | |
| − | <pre>
| + | |
| − | [GENERAL]
| + | |
| − | description = BLASTZ alignments for Oryza sativa
| + | |
| | | | |
| − | ===Sample Configuration Files=== | + | * Using the Generic Synteny Browser (GBrowse_syn) <ref name=PMID:20836076/> |
| − | # The synteny database
| + | |
| − | join = dbi:mysql:database=rice_synteny;host=localhost
| + | |
| | | | |
| − | # This option maps the relationship between the species data sources, names and descriptions
| + | ===Tutorials=== |
| − | # The value for "name" (the first column) is the symbolic name that gbrowse_syn users to identify each species.
| + | |
| − | # This value is also used in two other places in the gbrowse_syn configuration:
| + | |
| − | # the species name in the "examples" directive and the species name in the .aln file
| + | |
| − | # The value for "conf. file" is the basename of the corresponding gbrowse .conf files.
| + | |
| − | # This value is also used to identify the species configuration stanzas at the bottom of the configuration file.
| + | |
| | | | |
| − | # name conf. file Description
| + | *[[GBrowse_syn_Tutorial|Installing and configuring GBrowse_syn]] |
| − | source_map = rice rice_synteny "Domesic Rice (O. sativa)"
| + | |
| − | wild_rice wild_rice_synteny "Wild Rice"
| + | |
| | | | |
| − | tmpimages = /tmp/gbrowse2
| + | ===Presentations=== |
| − | imagewidth = 800
| + | |
| − | stylesheet = /gbrowse2/css/gbrowse_transparent.css
| + | |
| − | cache time = 1
| + | |
| | | | |
| − | config_extension = conf
| + | * [[:Media:GBrowse_syn_EBI2009.pdf|Challenges in Comparative Genome Browsing]] - Presented by [[User:Mckays|Sheldon McKay]] at the [http://www.ebi.ac.uk European Bioinformatics Institute], Hinxton, UK. |
| | + | * [[:Media:GBrowse_synSMBE2009.pdf|Comparative Genomics with GBrowse_syn]] - Presentation by [[User:Mckays|Sheldon McKay]] at the [http://ccg.biology.uiowa.edu/smbe/symposia.php?action=view&sym_ID=27 SMBE 2009 GMOD Workshop] on using [[GBrowse_syn]] for [[:Category:Comparative Genomics|comparative genomics]]. |
| | + | * [[GBrowse_syn PAG 2009 Workshop| GBrowse_syn at PAG]] - Presentation by [[User:Mckays|Sheldon McKay]] at the Plant and Animal Genomes meeting, San Diego, CA, USA. |
| | + | * [[Media:Gbrowse_syn.pdf|November 2007]] - [[User:Mckays|Sheldon McKay]]'s presentation on GBrowse_syn at the [[November 2007 GMOD Meeting#GBrowse_Syn|November 2007 GMOD Meeting]]. |
| | | | |
| − | # example searches to display
| + | ==Contacts and Mailing Lists== |
| − | examples = rice 3:16050173..16064974
| + | |
| − | wild_rice 3:1..400000
| + | |
| | | | |
| − | zoom levels = 5000 10000 25000 50000 100000 200000 400000
| + | Support is via the GBrowse mailing list: |
| | | | |
| − | # species-specific databases
| + | {| class="wikitable" |
| − | [rice_synteny] | + | | |
| − | tracks = EG
| + | ! Mailing List Link |
| − | color = blue
| + | ! Description |
| | + | ! Archive(s) |
| | + | |- |
| | + | ! rowspan="2" | [[GBrowse]] & [[GBrowse_syn]] |
| | + | | [https://lists.sourceforge.net/lists/listinfo/gmod-gbrowse gmod-gbrowse] |
| | + | | [[GBrowse]] and [[GBrowse_syn]] users and developers. |
| | + | | [http://dir.gmane.org/gmane.science.biology.gmod.gbrowse Gmane], [http://gmod.827538.n3.nabble.com/GBrowse-f815907.html Nabble (2010/05+)], [https://lists.sourceforge.net/lists/listinfo/gmod-gbrowse Sourceforge] |
| | + | |- |
| | + | | [https://lists.sourceforge.net/lists/listinfo/gmod-gbrowse-cmts gmod-gbrowse-cmts] |
| | + | | Code updates. |
| | + | | [http://sourceforge.net/mailarchive/forum.php?forum_name=gmod-gbrowse-cmts Sourceforge] |
| | + | |} |
| | | | |
| − | [wild_rice_synteny]
| |
| − | tracks = EG
| |
| − | color = red
| |
| − | </pre>
| |
| | | | |
| − | A sample species config file, <tt>rice_synteny.conf</tt>:
| + | ==GBrowse_syn in the wild== |
| − | <pre>
| + | |
| − | [GENERAL]
| + | |
| − | description = Domestic rice chromosome 3
| + | |
| − | db_adaptor = Bio::DB::SeqFeature::Store
| + | |
| − | db_args = -adaptor memory
| + | |
| − | -dir /var/www/gbrowse2/databases/gbrowse_syn/rice
| + | |
| | | | |
| | + | Public installations of GBrowse_syn: |
| | + | *[http://www.arabidopsis.org/cgi-bin/gbrowse_syn/arabidopsis/?name=Chr1%3A8367000..8370501 The Arabidopsis Information Resource] |
| | + | *[http://dev.wormbase.org/db/seq/gbrowse_syn/compara?search_src=Cele;name=X:1050001..1150000 WormBase] |
| | + | *[http://solgenomics.net/gbrowse2/bin/gbrowse_syn/sol3/ Sol Genomics] |
| | | | |
| − | # Web site configuration info
| + | ==GBrowse_syn Development== |
| − | tmpimages = /tmp/gbrowse2
| + | |
| | | | |
| − | [EG]
| |
| − | feature = gene:ensembl
| |
| − | glyph = gene
| |
| − | height = 10
| |
| − | bgcolor = peachpuff
| |
| − | fgcolor = hotpink
| |
| − | description = 0
| |
| − | label = 0
| |
| − | category = Transcripts
| |
| − | key = ensembl gene
| |
| − | balloon hover = Hello, my name is $name!
| |
| − |
| |
| − | </pre>
| |
| − | <div class="emphasisbox">
| |
| − | Note: the species databases are actually using the [[GFF3]] flat file, in-memory adapter
| |
| − | </div>
| |
| − |
| |
| − | ===Activating the Oryza Data Source===
| |
| − | 1) Make sure the temporary image directory specified in the config files exists and is world-writable
| |
| − | <div class="indent">
| |
| − | $ <span class="enter">sudo mkdir /var/www/tmp</span>
| |
| − | $ <span class="enter">sudo mkdir /var/www/tmp/gbrowse2</span>
| |
| − | $ <span class="enter">sudo chmod 777 /var/www/tmp/gbrowse2</span>
| |
| − | </div>
| |
| − |
| |
| − | 2) Renaming the configuration file
| |
| − | <div class="indent">
| |
| − | $ <span class="enter">cd /etc/gbrowse2/synteny</span>
| |
| − | $ <span class="enter">sudo mv oryza.synconf.disabled oryza.synconf</span>
| |
| − | </div>
| |
| − |
| |
| − | 3) Point your browser to http://localhost/cgi-bin/gb2/gbrowse_syn/oryza. You should see:
| |
| − | <div class="indent">
| |
| − | [[Image:We_made_it1.png|left|800px]]
| |
| − | <br clear=all>
| |
| − | </div>
| |
| − |
| |
| − | 4) Click on the first example, you should (eventually) see:
| |
| − | <div class="indent">
| |
| − | [[Image:We_made_it2.png|left|800px]]
| |
| − | <br clear=all>
| |
| − | </div>
| |
| − |
| |
| − | 5) Try out a few user interface features:
| |
| − | <div class="indent">
| |
| − | * mouse over one of the genes:
| |
| − | [[Image:bubble1.png]]<br clear=all>
| |
| − | * Click on one of the bold blue highlighted section titles. This takes you to a contextual help page on the GMOD wiki.
| |
| − | * Click and drag on the overview panel. This will trigger rubber band selection to recenter or resize the displayed image
| |
| − | </div>
| |
| − |
| |
| − | ==Speeding up the Browser==
| |
| − | You can speed up the image loading time by putting your species' [[GFF3]] data into relational MySQL databases.
| |
| − |
| |
| − | 1) Create a database for each of the GFF data files (<tt>rice.gff3</tt> and <tt>wild_rice.gff3</tt>).
| |
| − |
| |
| − | <div class="indent">
| |
| − | $ <span class="enter">mysql -uroot -pgmodamericas2011</span>
| |
| − |
| |
| − | mysql> <span class="enter">create database rice;</span>
| |
| − | Query OK, 1 row affected (0.00 sec)
| |
| − |
| |
| − | mysql> <span class="enter">create database wild_rice;</span>
| |
| − | Query OK, 1 row affected (0.00 sec)
| |
| − |
| |
| − | mysql> <span class="enter">quit</span>
| |
| − | </div>
| |
| − |
| |
| − | 2) Populate the databases using the [[GBrowse Install HOWTO#GFF3|Loading <tt>bp_seqfeature_load.pl</tt>]] (pre-installed as part of [[gmod:BioPerl|BioPerl]] with [[GBrowse]]). This will load the [[GFF3]] data into a MySQL relational database. Note the MySQL user will root-level privileges.
| |
| − |
| |
| − | <div class="indent">
| |
| − | $ <span class="enter">cd /var/www/gbrowse2/databases/gbrowse_syn/rice</span>
| |
| − | $ <span class="enter">bp_seqfeature_load.pl -u root -p gmodamericas2011 -d rice -c -f rice.gff3</span>
| |
| − | loading rice.gff3...
| |
| − | Building object tree...
| |
| − | 0.55s0s
| |
| − |
| |
| − | Loading bulk data into database... 0.73s
| |
| − | load time: 11.99s
| |
| − |
| |
| − | $ <span class="enter">cd ../wild_rice</span>
| |
| − | $ <span class="enter">bp_seqfeature_load.pl -u root -p gmodamericas2011 -d wild_rice -c -f wild_rice.gff3</span>
| |
| − | loading wild_rice.gff3...
| |
| − | Building object tree...
| |
| − | 0.55s7a
| |
| − |
| |
| − | Loading bulk data into database... 0.69s
| |
| − | load time: 12.02s
| |
| − | </div>
| |
| − |
| |
| − | 3) Modify the following stanza in the file <tt>rice_synteny.conf</tt> in cd /etc/gbrowse2/synteny/. This will convert your data source from a flat file database to a MySQL relational database.
| |
| − |
| |
| − | <div class="indent">
| |
| − | # from
| |
| − | db_args = -adaptor memory
| |
| − | -dir /var/www/html/gbrowse/databases/gbrowse_syn/rice
| |
| − |
| |
| − | # to
| |
| − | <span class="enter">db_args = -dsn dbi:mysql:rice</span>
| |
| − | </div>
| |
| − |
| |
| − | 4) repeat for <tt>wild_rice_synteny.conf</tt>
| |
| − |
| |
| − | =Using Non-alignment Data=
| |
| − | <div class="emphasisbox">
| |
| − | This example uses gene orthology-based synteny blocks* based created by [http://genome.sfu.ca/orthoclusterdb OrthoCluster] for three nematode species, <i>C. elegans</i>, <i>C. briggsae</i> and <i>P. pacificus</i>.
| |
| − | <p>
| |
| − | <font style="font-size:7pt">''*Data courtesy of Jack Chen and Ismael Vergera''</font>
| |
| − | </p>
| |
| − | </div>
| |
| − |
| |
| − | 1) Download and unpack the data archive file <tt>orthocluster.tar.gz</tt>.
| |
| − | <div class="indent">
| |
| − | $ <span class="enter">cd ~/Documents/Data/gbrowse_syn</span>
| |
| − | $ <span class="enter">rm orthocluster.tar.gz</span>
| |
| − | $ <span class="enter">wget ftp://ftp.gmod.org/pub/gmod/GBrowse_syn/orthocluster.tar.gz</span>
| |
| − | $ <span class="enter">tar zxf orthocluster.tar.gz</span>
| |
| − | </div>
| |
| − |
| |
| − | 2) Examine the contents of the <tt>ORTHOCLUSTER</tt> directory tree using the Unix <tt>tree</tt> command. It is not installed by default, so we will have to get it first.
| |
| − |
| |
| − | <div class="indent">
| |
| − | $ <span class="enter">sudo apt-get install tree</span>
| |
| − | [sudo] password for gmod:
| |
| − | Reading package lists... Done
| |
| − | Building dependency tree
| |
| − | Reading state information... Done
| |
| − | The following NEW packages will be installed:
| |
| − | tree
| |
| − | 0 upgraded, 1 newly installed, 0 to remove and 37 not upgraded.
| |
| − | Need to get 31.1kB of archives.
| |
| − | After this operation, 98.3kB of additional disk space will be used.
| |
| − | Get:1 http://us.archive.ubuntu.com karmic/universe tree 1.5.2.2-1 [31.1kB]
| |
| − | Fetched 31.1kB in 0s (37.0kB/s)
| |
| − | Selecting previously deselected package tree.
| |
| − | (Reading database ... 135915 files and directories currently installed.)
| |
| − | Unpacking tree (from .../tree_1.5.2.2-1_i386.deb) ...
| |
| − | Processing triggers for man-db ...
| |
| − | Setting up tree (1.5.2.2-1) ...
| |
| − |
| |
| − | Now we can use it
| |
| − |
| |
| − | $ <span class="enter">tree ORTHOCLUSTER/</span>
| |
| − | ORTHOCLUSTER/
| |
| − | |-- conf
| |
| − | | |-- bri.conf
| |
| − | | |-- ele.conf
| |
| − | | |-- orthocluster.synconf
| |
| − | | `-- ppa.conf
| |
| − | |-- gff
| |
| − | | |-- bri.gff
| |
| − | | |-- ele.gff
| |
| − | | `-- ppa.gff
| |
| − | `-- orthocluster.txt
| |
| − |
| |
| − | 2 directories, 8 files
| |
| − |
| |
| − | In the <tt>conf</tt> directory, there are configuration files for the joining database and each of the three species. They are similar in structure to the examples shown above, except that the database adapter {{CPAN|Bio::DB::GFF}} and a gene aggregator are used because the [[gmod:GFF2|GFF is version 2]]. For example:
| |
| − | <pre>
| |
| − | [GENERAL]
| |
| − | description = C. briggsae
| |
| − | db_adaptor = Bio::DB::GFF
| |
| − | db_args = -dsn dbi:mysql:bri
| |
| − |
| |
| − | # This is the GFF2 aggregator that assembles gene models
| |
| − | # from coding exon features with the same parent
| |
| − | aggregators = gene{coding_exon}
| |
| − |
| |
| − | </pre>
| |
| − |
| |
| − |
| |
| − | The <tt>gff</tt> directory contains gene annotations for each of the three species, derived from [http://www.wormbase.org WormBase] (release WS204). The files are in [[gmod:GFF2|GFF2]] format, which is why the {{CPAN|Bio::DB::GFF}} adapter is required. A sample is shown here:
| |
| − |
| |
| − | <pre>
| |
| − | ##gff-version 2
| |
| − | ##sequence-region I 1 15072421
| |
| − | ##sequence-region II 1 15279324
| |
| − | ##sequence-region III 1 13783685
| |
| − | ##sequence-region IV 1 17493784
| |
| − | ##sequence-region V 1 20924143
| |
| − | ##sequence-region X 1 17718854
| |
| − | I curated coding_exon 11641 11689 . + 0 CDS "Y74C9A.2"
| |
| − | I curated coding_exon 14951 15160 . + 2 CDS "Y74C9A.2"
| |
| − | I curated coding_exon 16473 16585 . + 2 CDS "Y74C9A.2"
| |
| − | I curated coding_exon 43733 43961 . + 0 CDS "Y74C9A.1"
| |
| − | I curated coding_exon 44030 44234 . + 2 CDS "Y74C9A.1"
| |
| − | I curated coding_exon 44281 44324 . + 1 CDS "Y74C9A.1"
| |
| − | I curated coding_exon 44372 44468 . + 2 CDS "Y74C9A.1"
| |
| − | I curated coding_exon 44521 44677 . + 1 CDS "Y74C9A.1"
| |
| − | I curated coding_exon 47472 47610 . + 0 CDS "Y48G1C.12"
| |
| − | I curated coding_exon 47696 47858 . + 2 CDS "Y48G1C.12"
| |
| − | I curated coding_exon 48348 48530 . + 1 CDS "Y48G1C.12"
| |
| − | I curated coding_exon 49251 49416 . + 1 CDS "Y48G1C.12"
| |
| − | </pre>
| |
| − |
| |
| − | The file <tt>orthocluster.txt</tt> contains the synteny data. The first few lines are shown below. The first 12 fields in each row specify information about the synteny block in each species and the series of numbers are orthologous gene coordinate pairs that are used for linking orthologs with grid-lines in the GBrowse_syn display. See 'Alignment Data' under [[#Further Reading|Further Reading]] below for more details of this loading format.
| |
| − | <pre>
| |
| − | bri chrI 176154 183558 + . ppa Ppa_Contig88 27212 30786 + . 176154 27212 177594 30786 182118 27212 183558 30786 | 30786 183558 27212 182118 30786 177594 27212 176154
| |
| − | bri chrI 778780 799223 + . ppa Ppa_Contig88 533454 542961 - . 778780 539924 786778 542961 789497 533454 799223 538726 | 538726 799223 533454 789497 542961 786778 539924 778780
| |
| − | bri chrI 986150 994698 + . ppa Ppa_Contig77 29481 45600 - . 986150 37055 989649 45600 991428 29481 994698 36608 | 36608 994698 29481 991428 45600 989649 37055 986150
| |
| − | bri chrI 1453793 1461931 + . ppa Ppa_Contig132 156183 165414 - . 1453793 163110 1456404 165414 1456712 160849 1457637 162712 1458361 160204 1459245 160815 1459468 159346 1459854 160000 1459962 156183 1461931 159022 | 159022 1461931 156183 1459962 160000 1459854 159346 1459468 160815 1459245 160204 1458361 162712 1457637 160849 1456712 165414 1456404 163110 1453793
| |
| − |
| |
| − | </pre></div>
| |
| − |
| |
| − |
| |
| − | 3) Set the <tt>$TMP</tt> environmental variable so that the database loading script knows where to put its temp files.
| |
| − |
| |
| − | <div class="indent">
| |
| − | $ <span class="enter">export TMP=/tmp</span>
| |
| − | </div>
| |
| − |
| |
| − | 4) Create and load a Bio::DB:GFF database for ''C. elegans'' (ele). Use screen so that we can get the time-consuming loading script started '''and then use <tt>Ctrl-A D</tt> to set the screen running in the background''' and move on to other steps.
| |
| − | <div class="indent">
| |
| − | $ <span class="enter">cd ORTHOCLUSTER/gff</span>
| |
| − | $ <span class="enter">mysql -uroot -pgmodamericas2011 -e 'create database ele'</span>
| |
| − | $ <span class="enter">screen bp_fast_load_gff.pl -u root -p gmodamericas2011 -d ele -c ele.gff</span>
| |
| − | </div>
| |
| − |
| |
| − | 5) Repeat step 4 for the other two species (bri and ppa).
| |
| − |
| |
| − |
| |
| − | 6) Create and load the alignment the alignment database. The <tt>gbrowse_syn_load_alignment_database.pl</tt> script is pre-installed with [[GBrowse]].
| |
| − |
| |
| − | <div class="indent">
| |
| − | $ <span class="enter">cd ..</span>
| |
| − | $ <span class="enter">mysql -uroot -pgmodamericas2011 -e 'create database orthocluster'</span>
| |
| − | $ <span class="enter">gbrowse_syn_load_alignment_database.pl -u root -p gmodamericas2011 -d orthocluster -c -v orthocluster.txt</span>
| |
| − | </div>
| |
| − |
| |
| − | 7) Copy the configuration files to the required location
| |
| − |
| |
| − | <div class="indent">
| |
| − | $ <span class="enter">cd conf</span>
| |
| − | $ <span class="enter">sudo cp *conf /etc/gbrowse2/synteny</span>
| |
| − | </div>
| |
| | | | |
| − | 8) Go back to your browser and reload the rice page. There should now be a second data source in a pull-down menu.
| + | See the [[Talk:GBrowse_syn|discussion page]] for notes on further GBrowse_syn development. |
| − | <br>
| + | |
| − | :[[Image:pulldown1.png]]<br clear=all>
| + | |
| | | | |
| − | 9) Select the other data source and start browsing!
| |
| − | <br>
| |
| − | :[[Image:etfinit.png|border|left|700px]]<br clear=all>
| |
| | | | |
| − | =Further Reading= | + | ==See also== |
| − | ==A Note on Whole Genome Alignments== | + | |
| − | The focus of the section of the course is on dealing with alignment or synteny data and using [[GBrowse_syn]]. However, how to generate whole genome alignments, identify orthologous regions, etc, are the subject of considerable interest, so some background reading is listed below:
| + | |
| − | *<span class="pops">[http://www.eecs.berkeley.edu/Pubs/TechRpts/2006/EECS-2006-104.html Primer on Hierarchical Genome Alignment Strategies]</span>
| + | |
| − | *<span class="pops">[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2577869 article on PECAN and ENREDO]</span>
| + | |
| − | *<span class="pops">[http://www.ebi.ac.uk/~bjp/pecan/ all about PECAN]</span>
| + | |
| − | * The gene annotations for each species are in [[GFF]] files.
| + | |
| − | * The alignment data are in a <span class="pops">[[gmod:GBrowse_syn Database#Clustal alignment format|constrained CLUSTALW format]]</span> (They were not generated by the program CLUSTALW, which is not necessarily suitable for whole genome alignments)
| + | |
| − | * There are processing steps for the alignment data but it is very computationally intensive and we will load pre-processed data to get a head start.
| + | |
| | | | |
| − | ==Documentation==
| + | The focus of this documentation is the GBrowse_syn application. However, the generation of whole genome alignments and identification of orthologous regions are the subject of considerable interest, so some background reading is listed below: |
| − | There is detailed documentation on the GMOD wiki for how to install, configure and use GBrowse_syn. To get started, browse these pages:
| + | *[http://www.eecs.berkeley.edu/Pubs/TechRpts/2006/EECS-2006-104.html Primer on Hierarchical Genome Alignment Strategies] |
| − | *<span class="pops">[[gmod:GBrowse_syn|GBrowse_syn overview]]</span> | + | *[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2577869/ article on PECAN and ENREDO] |
| − | *<span class="pops">[[gmod:GBrowse_syn#Installation|Installation]]</span>
| + | *[http://www.ebi.ac.uk/~bjp/pecan/ all about PECAN] |
| − | *<span class="pops">[[gmod:GBrowse_syn Configuration|Configuration]]</span>
| + | *[http://www.ensembl.org/info/website/archives/index.html Information about EnsEMBL's compara pipeline] |
| − | *<span class="pops">[[gmod:GBrowse_syn Database|Alignment Data]]</span> | + | |
| − | *<span class="pops">[[gmod:GBrowse_syn Help|The user interface]]</span> | + | |
| − | *<span class="pops">[[gmod:GBrowse_syn#Presentations_and_Workshops|Presentations and workshops]]</span> | + | |
| | | | |
| − | = Evaluation = | + | ==More on GBrowse_syn== |
| | | | |
| | + | See [[:Category:GBrowse_syn]] |
| | + | The [[:Image:GBrowse_syn_logo.png|GBrowse_syn logo]] was created by [mailto:NextLevelDesignStudios@gmail.com Darek Lakey], a participant in the [[Spring 2010 Logo Program]], while a design student at [http://www.linnbenton.edu Linn-Benton Community College]. |
| | | | |
| − | {{Feedback}}
| |
| | | | |
| − | {{NextSession|Galaxy|Galaxy}}
| + | <references /> |
| | | | |
| − | <hr>
| + | [[Category:GBrowse syn]] |
| − | <div style="float:right">[[Image:iPlant.png|250px]]</div>
| + | [[Category:GMOD Components]] |
| | + | [[Category:GMOD Developers]] |
| | + | [[Category:GBrowse]] |
| | + | [[Category:Comparative Genomics]] |
| | + | [[Category:WormBase]] |
The overall look of Gbrowse_syn resembles GBrowse but has some key differences to accomodate the more complex comparative genome data (see the screenshot above).
GBrowse_syn uses a central "reference species" panel, with inset panels above and below for two or more aligned species. There is no upper limit to the number of species that can be displayed.
The focus of this documentation is the GBrowse_syn application. However, the generation of whole genome alignments and identification of orthologous regions are the subject of considerable interest, so some background reading is listed below: