Difference between revisions of "GBrowse"
m (→Description: Droped DNA hyperlink) |
|||
| Line 20: | Line 20: | ||
* Third-party feature loading. | * Third-party feature loading. | ||
* Customizable plug-in architecture (e.g. run [[wp:BLAST|BLAST]], dump & import many formats, find oligonucleotides, design primers, create restriction maps, edit features) | * Customizable plug-in architecture (e.g. run [[wp:BLAST|BLAST]], dump & import many formats, find oligonucleotides, design primers, create restriction maps, edit features) | ||
| + | |||
| + | ==Installation== | ||
| + | |||
| + | * The network installer script: {{CVS|Generic-Genome-Browser/bin/gbrowse_netinstall.pl}} | ||
| + | ** You will need to have Perl 5.8.6 or higher, and the Apache web server installed. See the step-by-step instructions below before running the installer for discussion of prerequisites needed. Once these are installed, save the above link to your home directory as gbrowse_netinstall.pl, and then run "perl gbrowse_netinstall.pl". On Windows platforms, you will need to be logged in as a user with administrative privileges. On Mac OSX, Linux or Solaris platforms, you will need to be logged in as root, or to run the command using "sudo" (''sudo perl gbrowse_netinstall.pl''). | ||
| + | |||
| + | The install script has a few useful options: | ||
| + | <pre> | ||
| + | -h|--help : Show this message | ||
| + | -d|--dev : Use the developement version of both GBrowse and bioperl from CVS | ||
| + | --bioperl_dev : Use the development version of BioPerl from CVS | ||
| + | --gbrowse_dev : Use the development version of GBrowse from CVS | ||
| + | -b|--build_param_str : Use this string to predefine Makefile.PL parameters | ||
| + | such as CONF or PREFIX for GBrowse installation | ||
| + | </pre> | ||
| + | |||
| + | Note that the -d option does not work on Windows, as there is no system support for cvs on Windows. We are working on a way around this problem. | ||
| + | |||
| + | So, for example, if you want to install the most recent developments in GBrowse and BioPerl, you can do this: | ||
| + | |||
| + | sudo perl gbrowse_netinstall.pl -d | ||
| + | |||
| + | This option is useful for getting new options just added to GBrowse (for example, the Balloon pop up windows added before the release of GBrowse 1.69) or for getting a new BioPerl that has recent bugs fixed. | ||
| + | |||
| + | * Detailed step-by-step install documentation | ||
| + | ** [[GBrowse_MacOSX_HOWTO|Install on MacOSX]] | ||
| + | ** [[GBrowse_Windows_HOWTO|Install on Windows]] | ||
| + | ** [[GBrowse_Ubuntu_HOWTO|Install on Ubuntu and other Debian-based systems]] | ||
| + | ** [[GBrowse_RPM_HOWTO|Install on Fedora Core and other RPM-based systems]] | ||
| + | ** [[GBrowse_Gentoo_HOWTO|Install on Gentoo Linux system]] | ||
| + | ** [[GBrowse_Install_HOWTO|Source Code Install (for other Linux systems)]] | ||
==Documentation== | ==Documentation== | ||
| Line 35: | Line 66: | ||
* {{CVS|Generic-Genome-Browser/docs/pod/BIOSQL_ADAPTER_HOWTO.pod}} | * {{CVS|Generic-Genome-Browser/docs/pod/BIOSQL_ADAPTER_HOWTO.pod}} | ||
| − | * {{CVS|Generic-Genome-Browser/docs/pod/GENBANK_HOWTO.pod}} | + | * {{CVS|Generic-Genome-Browser/docs/pod/GENBANK_HOWTO.pod}} |
* {{CVS|Generic-Genome-Browser/docs/pod/PLUGINS_HOWTO.pod}} | * {{CVS|Generic-Genome-Browser/docs/pod/PLUGINS_HOWTO.pod}} | ||
| − | * {{CVS|Generic-Genome-Browser/docs/pod/CONFIGURE_HOWTO.pod}} | + | * {{CVS|Generic-Genome-Browser/docs/pod/CONFIGURE_HOWTO.pod}} |
| − | * {{CVS|Generic-Genome-Browser/docs/pod/INSTALL.MacOSX.pod}} | + | * {{CVS|Generic-Genome-Browser/docs/pod/INSTALL.MacOSX.pod}} |
| − | * {{CVS|Generic-Genome-Browser/docs/pod/DAS_HOWTO.pod}} | + | * {{CVS|Generic-Genome-Browser/docs/pod/DAS_HOWTO.pod}} |
| − | * {{CVS|Generic-Genome-Browser/docs/pod/INSTALL.pod}} | + | * {{CVS|Generic-Genome-Browser/docs/pod/INSTALL.pod}} |
* {{CVS|Generic-Genome-Browser/docs/pod/README-chado.pod}} | * {{CVS|Generic-Genome-Browser/docs/pod/README-chado.pod}} | ||
| − | * {{CVS|Generic-Genome-Browser/docs/pod/FAQ.pod}} | + | * {{CVS|Generic-Genome-Browser/docs/pod/FAQ.pod}} |
| − | * {{CVS|Generic-Genome-Browser/docs/pod/MAKE_IMAGES_HOWTO.pod}} | + | * {{CVS|Generic-Genome-Browser/docs/pod/MAKE_IMAGES_HOWTO.pod}} |
* {{CVS|Generic-Genome-Browser/docs/pod/README-gff-files.pod}} | * {{CVS|Generic-Genome-Browser/docs/pod/README-gff-files.pod}} | ||
| − | * {{CVS|Generic-Genome-Browser/docs/pod/GBROWSE_IMG.pod}} | + | * {{CVS|Generic-Genome-Browser/docs/pod/GBROWSE_IMG.pod}} |
| − | * {{CVS|Generic-Genome-Browser/docs/pod/ORACLE_AND_POSTGRESQL.pod}} | + | * {{CVS|Generic-Genome-Browser/docs/pod/ORACLE_AND_POSTGRESQL.pod}} |
* {{CVS|Generic-Genome-Browser/docs/pod/README-lucegene.pod}} | * {{CVS|Generic-Genome-Browser/docs/pod/README-lucegene.pod}} | ||
| Line 73: | Line 104: | ||
You can also browse the [http://gmod.cvs.sourceforge.net/gmod/Generic-Genome-Browser GBrowse CVS]. | You can also browse the [http://gmod.cvs.sourceforge.net/gmod/Generic-Genome-Browser GBrowse CVS]. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
== About Databases == | == About Databases == | ||
| Line 116: | Line 117: | ||
==Contacts== | ==Contacts== | ||
| − | Please report bugs to the SourceForge [http://sourceforge.net/tracker/?func=add&group_id=27707&atid=391291 Bug Tracker]. | + | Please report bugs to the SourceForge [http://sourceforge.net/tracker/?func=add&group_id=27707&atid=391291 Bug Tracker]. |
Please send questions to the [https://lists.sourceforge.net/lists/listinfo/gmod-gbrowse GBrowse mailing list]. | Please send questions to the [https://lists.sourceforge.net/lists/listinfo/gmod-gbrowse GBrowse mailing list]. | ||
Revision as of 23:00, 5 February 2008
Contents
Description
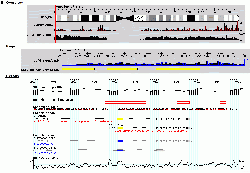
GBrowse[1] is the most popular viewer in GMOD. For a list of GBrowse and GMOD installations see the GMOD Users page. For a demo of its features, try the WormBase, FlyBase, or Human Genome Segmental Duplication Database web sites.
The Generic Genome Browser is a combination of database and interactive Web page for manipulating and displaying annotations on genomes. Some of its features:
- Simultaneous bird's eye and detailed views of the genome.
- Scroll, zoom, center.
- Use a variety of premade glyphs (see the BioPerl Glyph documentation for a list) or create your own.
- Attach arbitrary URLs to any annotation.
- Order and appearance of tracks are customizable by administrator and end-user.
- Search by annotation ID, name, or comment.
- Supports third party annotation using GFF formats.
- Settings persist across sessions.
- DNA and GFF dumps.
- Connectivity to different databases, including BioSQL and Chado.
- Multi-language support.
- Third-party feature loading.
- Customizable plug-in architecture (e.g. run BLAST, dump & import many formats, find oligonucleotides, design primers, create restriction maps, edit features)
Installation
- The network installer script: Generic-Genome-Browser/bin/gbrowse_netinstall.pl
- You will need to have Perl 5.8.6 or higher, and the Apache web server installed. See the step-by-step instructions below before running the installer for discussion of prerequisites needed. Once these are installed, save the above link to your home directory as gbrowse_netinstall.pl, and then run "perl gbrowse_netinstall.pl". On Windows platforms, you will need to be logged in as a user with administrative privileges. On Mac OSX, Linux or Solaris platforms, you will need to be logged in as root, or to run the command using "sudo" (sudo perl gbrowse_netinstall.pl).
The install script has a few useful options:
-h|--help : Show this message
-d|--dev : Use the developement version of both GBrowse and bioperl from CVS
--bioperl_dev : Use the development version of BioPerl from CVS
--gbrowse_dev : Use the development version of GBrowse from CVS
-b|--build_param_str : Use this string to predefine Makefile.PL parameters
such as CONF or PREFIX for GBrowse installation
Note that the -d option does not work on Windows, as there is no system support for cvs on Windows. We are working on a way around this problem.
So, for example, if you want to install the most recent developments in GBrowse and BioPerl, you can do this:
sudo perl gbrowse_netinstall.pl -d
This option is useful for getting new options just added to GBrowse (for example, the Balloon pop up windows added before the release of GBrowse 1.69) or for getting a new BioPerl that has recent bugs fixed.
- Detailed step-by-step install documentation
Documentation
On-line documentation
- GBrowse Tutorial
- GBrowse installation HOWTO
- GBrowse configuration HOWTO
- Generic-Genome-Browser/htdocs/general_help.html
- Generic-Genome-Browser/htdocs/annotation_help.html
- GBrowse Popup Balloons
- GBrowse FAQ
POD documentation
There are many useful POD documents included with the distribution. These are converted to HTML files when you install the package, and can be found in /gbrowse/docs/pod:
- Generic-Genome-Browser/docs/pod/BIOSQL_ADAPTER_HOWTO.pod
- Generic-Genome-Browser/docs/pod/GENBANK_HOWTO.pod
- Generic-Genome-Browser/docs/pod/PLUGINS_HOWTO.pod
- Generic-Genome-Browser/docs/pod/CONFIGURE_HOWTO.pod
- Generic-Genome-Browser/docs/pod/INSTALL.MacOSX.pod
- Generic-Genome-Browser/docs/pod/DAS_HOWTO.pod
- Generic-Genome-Browser/docs/pod/INSTALL.pod
- Generic-Genome-Browser/docs/pod/README-chado.pod
- Generic-Genome-Browser/docs/pod/FAQ.pod
- Generic-Genome-Browser/docs/pod/MAKE_IMAGES_HOWTO.pod
- Generic-Genome-Browser/docs/pod/README-gff-files.pod
- Generic-Genome-Browser/docs/pod/GBROWSE_IMG.pod
- Generic-Genome-Browser/docs/pod/ORACLE_AND_POSTGRESQL.pod
- Generic-Genome-Browser/docs/pod/README-lucegene.pod
Since these are in Perl POD format these files may contain formatting code when viewed in a Web browser.
Downloads
Source Code Download (tar.gz file)
Download the source from the SourceForge download page.
Net-based Installer Script
The net installer script, located at the GBrowse CVS repository will automatically download and install GBrowse and its Perl libraries for you. See Installation for details on using this script.
CVS
There are many new features in the current development version which have not been released yet. To get the latest version, please use anonymous CVS. The recommended branch to use is stable which is more stable than HEAD. Here is the recipe:
% cvs -d :pserver:anonymous@gmod.cvs.sourceforge.net:/cvsroot/gmod login CVS password: <hit return> % cvs -d :pserver:anonymous@gmod.cvs.sourceforge.net:/cvsroot/gmod co -r stable Generic-Genome-Browser
Once you have successfully checked out the Generic-Genome-Browser distribution, you can simply perform a cvs update inside the directory to get recent changes.
You can also browse the GBrowse CVS.
About Databases
GBrowse has a flexible adaptor system for running off various types of database. Standard adaptors include:
- Flat file adaptors (in-memory, indexed) -- put your annotations in a directory and go!
- Relational database adaptors -- Chado, BioSQL.
- Network adaptors -- read annotations from GenBank, UCSC or Ensembl.
- GFF databases -- Bio::DB::GFF, Bio::DB::SeqFeature
Contacts
Please report bugs to the SourceForge Bug Tracker.
Please send questions to the GBrowse mailing list.
References
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedPMID:12368253