NOTE: We are working on migrating this site away from MediaWiki, so editing pages will be disabled for now.
Difference between revisions of "GBrowse syn"
m (Merging Synteny Category with Comparative Genomics.) |
|||
| Line 9: | Line 9: | ||
*It comes as part of the GBrowse distribution, version 1.69 and later ([[GBrowse#On-line_documentation|GBrowse installation info...]]). | *It comes as part of the GBrowse distribution, version 1.69 and later ([[GBrowse#On-line_documentation|GBrowse installation info...]]). | ||
*It differs in that databases and configuration for individual species are linked together via a central configuration file and a joining database that contains reciprocal alignments between all species represented in the browser. | *It differs in that databases and configuration for individual species are linked together via a central configuration file and a joining database that contains reciprocal alignments between all species represented in the browser. | ||
| − | |||
| − | |||
| − | |||
| − | |||
==Configuration== | ==Configuration== | ||
===Configuration files=== | ===Configuration files=== | ||
| Line 33: | Line 29: | ||
===The Alignment database=== | ===The Alignment database=== | ||
| + | '''Example alignment database''' | ||
| + | To get an idea of how alignment database is built, the files and scripts used to load the synteny browser at [http://dev.wormbase.org/db/seq/gbrowse_syn WormBase] can be obtained from the [ftp://ftp.wormbase.org/pub/wormbase/misc/synteny_browser WormBase ftp site]. | ||
Revision as of 20:37, 12 August 2008
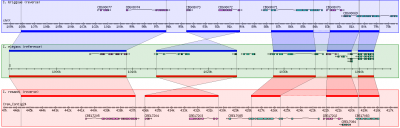
GBrowse_syn, or the Generic Synteny Browser, is a GBrowse-based synteny browser designed to display multiple genomes, with a central reference species compared to two or more additional species. It can be used to view multiple sequence alignment data, synteny or co-linearity data from other sources against genome annotations provided by GBrowse. GBrowse_syn is included with the standard GBrowse package (version 1.69 and later). Working examples can be seen at TAIR, WormBase, and SGN.
Contents
Documentation
Installation
- GBrowse_syn uses much of the same infrastructure as GBrowse, in terms of species databases, configuration files, perl libraries, etc.
- It comes as part of the GBrowse distribution, version 1.69 and later (GBrowse installation info...).
- It differs in that databases and configuration for individual species are linked together via a central configuration file and a joining database that contains reciprocal alignments between all species represented in the browser.
Configuration
Configuration files
The main configuration file specifies the alignment database, the species to be included and their corresponding configuration files.
- This file ends with the extension ".synconf".
- An example of the config file for the WormBase synteny browser can be seen here.
The species configuration file contains information used by each species-specific database.
- This file has the extension ".syn".
- This file is identical in structure to a normal GBrowse configuration file.
- To avoid confusing GBrowse, select names for your GBrowse_syn configuration files that are not similar to the names or regular GBrowse configuration files.
- An example of a species config file for the WormBase synteny browser can be seen here
The Alignment database
Example alignment database To get an idea of how alignment database is built, the files and scripts used to load the synteny browser at WormBase can be obtained from the WormBase ftp site.
Presentations
- November 2007 - Sheldon McKay's presentation on GBrowse_syn at the November 2007 GMOD Meeting.