Difference between revisions of "Overview"
m (→What is Chado?) |
m (→What is Chado?) |
||
| Line 88: | Line 88: | ||
ontologies to describe and organize our data. | ontologies to describe and organize our data. | ||
| − | Chado's focus is broader. | + | Chado's focus is broader. Its tables are broken down into groups called |
''modules'' and the modules are the following: | ''modules'' and the modules are the following: | ||
Revision as of 15:40, 8 March 2007
Contents
Introduction
With the amount of technical documentation available for GMOD the casual observer would be forgiven if they concluded that GMOD was a project about software. But it's not, GMOD has been created for biologists and in the real world it's used by biologists. However, the creators of GMOD are mostly not practicing biologists and the look and the feel of most GMOD documentation reflects this. What we will attempt to do is discuss GMOD from the researchers' perspective. This does not simply mean describe what the software does. If you look, for example, at a typical GBrowse page like this GBrowse view of human chromosome 7 you'll understand immediately what GBrowse is built to do, and a few more minutes of clicking and scrolling will reveal all sorts of useful ways to display and query the data. A modern biologist knows a great deal about bioinformatics functionality already. What we're more concerned with here are the practical details. Like given the data I have what database should I use? or do I even need a database? Or how hard is this going to be?
In our experience we find that most biologists want to focus on the science. They may have little knowledge of programming languages or databases, and only passing interest in the IT minutiae. They have deep knowledge of their own data, needless to say, and know how data like their own can be viewed and analyzed. What they want to know is how to create their own useful set of tools for their own data in as efficient a way as possible. And when this tool set is created they want to rest assured that their platform can be easily maintained in an environment where resources may be limited. We will attempt to address these sorts of questions.
By the way, the word we used here refers to the GMOD Help Desk. The Help Desk is a good resource for biologists who want to learn more about GMOD, for whatever reason. Feel free to email us at wg-emod@nescent.org.
What is a GMOD?
GMOD is a collection of interconnected applications and databases that biologists use as repositories and as tools. That connectivity is really the key here. Bioinformatic applications and databases are produced at a steady rate and this output is described each month in a number of different journals. There's no lack of tools, but many of these tools will be little used since the typical prospective user may not have the resources or expertise required to install the tool and connect it, in some way, to the data in hand. What is generally lacking is a concerted effort to produce tools and databases that will work together.
GMOD also describes a community. Many of the pieces of GMOD, or components, are mature software with many human-years of software development behind them. This amount of effort focussed on design, development, and testing has not occurred simply because someone wanted to code. The demand for software like this has been strong since genome sequence started to appear and many of the first genome databases used GMOD components. So GMOD describes this diverse group of software developers, scientists, and laboratories that use or improve these software components every day.
GMOD is also that specific thing that's installed on your computer. It may be the private viewer to your latest data that a student set up over the weekend. It may a Tb-scale database and suite of public Web applications developed over many years at a central laboratory. It may a database of experimental data that's accessible by script, or it may the annotation tool that you use to describe your favorite genome. Now, by describing this variety are we assuring you that whatever you want to do is possible within GMOD? No. The biologists lead and the software developers follow, not the other way around. So you may find that your predicament is not addressed, or is only partially addressed, by what's available in GMOD. You have an option here, which is to do something about this. First, contact the GMOD Help Desk or one of the main mailing lists like gmod-schema to make sure that your understanding of the available GMOD resources is correct. When you're in touch with some knowledgable person try to get a sense of what the solution might be, or its degree of difficulty. It may be that your solution may entail something simple, or it may be that a project may have to be created, complete with partnerships and grants. Be assured that the GMOD participants are very interested in seeing GMOD take off in new directions.
Is It Just for Model Organisms?
At first GMOD stood for Generic Model Organism Database, this was back in the days when there were a handful of model organisms and it appeared that obtaining the genomic sequence of an organism was a prohibitively expensive proposition, taking months or years to accomplish. Now there are hundreds of such sequences, with thousands conceivable. However, few of the scientists studying organisms with sequence consider their organism a model, in this first sense of the word.
This is a problem for the acronym since any organism with any kind of sequence associated with it is a good candidate as a subject for a GMOD database. So, for example, there are GMOD databases with just protein sequence in them like the S. cerevisiae Proteome Browser. There are GMOD databases with EST sequence only, such as the Cattle EST Gene Family Database. There are GMOD databases that are concerned primarily with gene expression, such as the Emiliania huxleyi Serial Analysis of Gene Expression database. We even find GMOD databases dedicated to collections of RNA sequence like the Leishmania tarentolae RNA Editing database. We have also heard of GMOD databases for things like oligonucleotides and plasmids. See GMOD Users for a list of other examples. That list of GMOD databases demonstrates that GMOD is widely used, with all sorts of organisms represented, and that these databases can hold sequences of any kind.
Some clever scientists have proposed that we just drop Model from the name and "re-brand" ourselves. GMOD thought about it and decided it may cause more problems than it solves. Instead, think of the M as standing for My or Many or Myriad.
What is GBrowse?
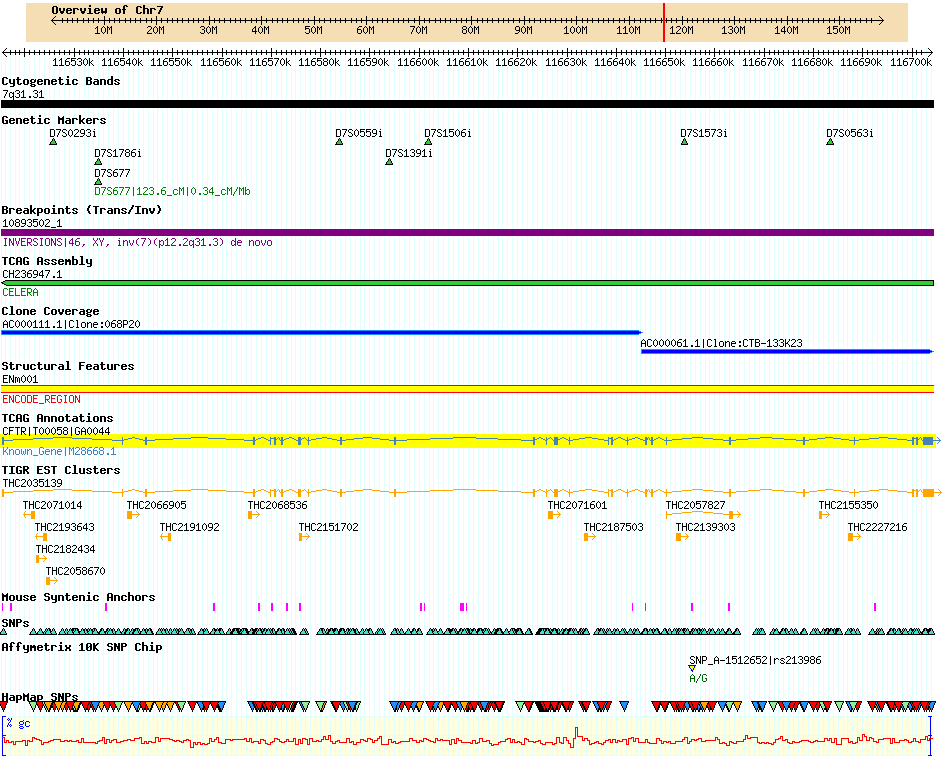
GBrowse is short for Genome Browser, or Generic Genome Browser. GBrowse is probably GMOD's most popular component and almost all of the databases listed in GMOD Users use GBrowse. It is fairly easy to install, only basic command-line familiarity is required. Do not be misled by the simplicity of the installation though: the explanation for GBrowse's popularity is that is a supremely capable browser. The picture below is a partial screenshot taken from the Human chromosome 7 database at TCAG. A bit of jargon: the rows, each depicting one sort of data, are called tracks and tracks are populated by one or more glyphs, plus text.
As of this writing GBrowse comes with some 75 different glyphs, including pie charts, dot plots, histograms, and X-Y plots suitable for quantitative data, as well as the expected array of glyphs that describe sequences and sequence annotation. It is also highly configurable, meaning you can do quite a bit of customization of the glyphs, you can link glyphs to URLs of your choice, you can internationalize the application to display different languages, you can connect and retrieve data from any database, and more. This sort of work generally requires either modifying GBrowse's configuration files or adding your own Perl code, the language that GBrowse is written in. Any customization requiring work in Perl should be considered routine coding, not difficult, and the explanation for this is that GBrowse is built to be customized.
What is Chado?
Chado is one of the relational databases that are used in GMOD, the other being BioSQL. Relational databases are today's tool of choice when faced with the problem of storing complex or multifaceted data, assuming that the data is, or can be, broken down into ever smaller bits of data. All atomized data will end up in one field, analogous to the way that data can be organized as columns in a spreadsheet. Fields describing one sort of thing are organized together into tables (but database designers do not talk about things, rather entities). For example, a relational database may have a table called gene with gene.name and gene.geneid' fields and a protein table with protein.name, protein.proteinid and protein.sequence fields.
The picture above shows these two tables. The relation between the
tables is the shared geneid - we add the geneid field to the protein
table to indicate that the CFTR_1 protein record relates
back to a specific gene in the gene table.
For a given collection of data, genomic sequence and annotation for example, there will be more than one way to represent the data relationally. A given relational design, essentially tables and fields, is called a schema (think of the schema as a blueprint, empty, and the schema populated with data as the database). Both Chado and BioSQL can store genomic data for example, but they do it differently. The details of how one designs relational schemas is not relevant here but can say that the designer may think about some of these general concerns:
- The degree of data abstraction, which is related to a database concept called normalization and to the flexibility of the schema
- The legibility of the schema, which has to do with the ease of using it
- The breadth of the schema, in terms of the data types it could contain
From the scientific perspective one can ask related questions: how flexible is a given schema? Can it handle my data now and in the future? Will using a given schema be easier or harder to use than some other schema?
This last question relates mostly to the degree of abstraction of the schema, not to the actual programming languages used. All of today's relational databases are created and loaded and queried using one language, SQL. Essentially what the programmers do is use their chosen language (Perl, Java, Python, etc.) to execute SQL and they all do this equally well.
So it will all really come back to you and your data, not technical details. In GMOD's case the choices are fairly clear. BioSQL is quite focussed, it's concerned with:
- Sequence
- Sequence annotation
- Phylogeny
It is also a thoroughly modern schema in that it uses OBO-style ontologies such as GO, the Gene Ontology. This is a requirement nowadays given the ability of ontologies to describe and organize our data.
Chado's focus is broader. Its tables are broken down into groups called modules and the modules are the following:
What is GMODWeb?
What is Bioperl?
What is Modware?
More Information
Resources
People
Hardware
Operating System
Software
Licenses
Case Studies
[Category:Needs Editing]]