Difference between revisions of "JBrowse"
From GMOD
MitchSkinner (Talk | contribs) m |
MitchSkinner (Talk | contribs) (change to new demo URL) |
||
| Line 1: | Line 1: | ||
{{SummerSchoolBoxBoth|JBrowse}} | {{SummerSchoolBoxBoth|JBrowse}} | ||
JBrowse is a genome browser with almost entirely [[Glossary#AJAX|AJAX]]-based interfaces. JBrowse renders most tracks using client side JavaScript and [[Glossary#JSON|JSON]] as its data transfer format. JBrowse is currently under development by the Ian Holmes Lab at UC Berkeley. It is expected to have a production release in 2009. Most development has been done by Mitch Skinner. JBrowse was formerly known as ''GBrowse 3''. However, this name was changed to JBrowse to avoid confusion: JBrowse is an alternative to [[GBrowse]], not a successor. | JBrowse is a genome browser with almost entirely [[Glossary#AJAX|AJAX]]-based interfaces. JBrowse renders most tracks using client side JavaScript and [[Glossary#JSON|JSON]] as its data transfer format. JBrowse is currently under development by the Ian Holmes Lab at UC Berkeley. It is expected to have a production release in 2009. Most development has been done by Mitch Skinner. JBrowse was formerly known as ''GBrowse 3''. However, this name was changed to JBrowse to avoid confusion: JBrowse is an alternative to [[GBrowse]], not a successor. | ||
| + | |||
| + | == Demo == | ||
| + | |||
| + | [http://jbrowse.org/genomes/dmel/ D. melanogaster] | ||
== Requirements == | == Requirements == | ||
| Line 22: | Line 26: | ||
[http://github.com/jbrowse From GitHub]. | [http://github.com/jbrowse From GitHub]. | ||
| − | == | + | == Screenshots == |
| − | + | ||
| − | + | ||
[[Image:JBrowseShot1.jpg|border]] | [[Image:JBrowseShot1.jpg|border]] | ||
Revision as of 02:12, 25 March 2009
JBrowse is a genome browser with almost entirely AJAX-based interfaces. JBrowse renders most tracks using client side JavaScript and JSON as its data transfer format. JBrowse is currently under development by the Ian Holmes Lab at UC Berkeley. It is expected to have a production release in 2009. Most development has been done by Mitch Skinner. JBrowse was formerly known as GBrowse 3. However, this name was changed to JBrowse to avoid confusion: JBrowse is an alternative to GBrowse, not a successor.
Contents
Demo
Requirements
- From CPAN:
- BioPerl 1.6
- JSON
- JSON::XS (optional, for speed)
- libpng
Documentation
- JBrowse Home Page
- JBrowse Quick Tutorial - JBrowse administration.
Presentations
- Image:GBrowse3GMOD2008.pdf - Presentation by Ian Holmes at July 2008 GMOD Meeting. See the talk's summary for more.
- JBrowse - Presentation by Mitch Skinner at the January 2009 GMOD Meeting. See the talk's summary for more.
Downloads
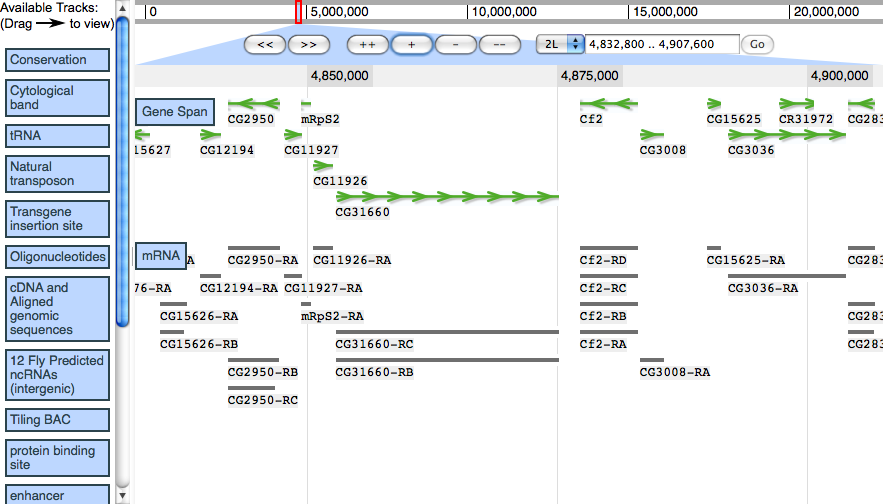
Screenshots
Demo screenshot from 2009/02/21, showing region of Drosophila melanogaster 2L.
Mailing List
- gmod-ajax@lists.sourceforge.net: Discussion of development, installation problems, etc.
- Subscribe to gmod-ajax
- gmod-ajax archives