NOTE: We are working on migrating this site away from MediaWiki, so editing pages will be disabled for now.
Difference between revisions of "GBrowse syn Database"
From GMOD
(→Database Loading Scripts) |
(→Database Loading Scripts) |
||
| Line 38: | Line 38: | ||
gunzip my_huge_clustal_file.aln.gz | perl split_clustal.pl /path/to/smaller_alignment_files | gunzip my_huge_clustal_file.aln.gz | perl split_clustal.pl /path/to/smaller_alignment_files | ||
| − | 2) parse the alignments using the script [http://gmod.cvs.sourceforge.net/viewvc/*checkout*/gmod/Generic-Genome-Browser/bin/gbrowse_syn/clustal2hit.pl?pathrev=stable clustal2hits.pl] | + | 2) parse the alignments using the script <span class="pops">[http://gmod.cvs.sourceforge.net/viewvc/*checkout*/gmod/Generic-Genome-Browser/bin/gbrowse_syn/clustal2hit.pl?pathrev=stable clustal2hits.pl]</span> |
| + | |||
| + | perl clustal2hit.pl /path/to/smaller_alignment_files/*.aln |gzip -c >processed_alignments.txt.gz | ||
| + | |||
| + | The output of this script is an ''ad hoc'' intermediate format that encodes the alignment coordinates plus 1:1 mapping of coordinates within the alignment (to facilitate accurate grid-lines in GBrowse_syn) | ||
| + | |||
| + | <pre> | ||
| + | #species1 seqid1 start1 end1 strand1 reserved species2 seqid2 start2 end2 strand2 reserved | ||
| + | c_briggsae chrI 1583997 1590364 + . c_remanei Crem_Contig24 631879 634679 - . \ | ||
| + | # pos1-1 pos1-2 ... posn-1 posn-2 | # pos1-2 pos1-1 ... posn-2 posn-1 | ||
| + | 1584000 634676 1584100 634584 (truncated for display...) | 631900 1590333 632000 1590233 (truncated for display...) | ||
| + | </pre> | ||
| + | |||
| + | |||
| + | 3) create a mysql database | ||
| + | |||
| + | mysql -uroot -ppassword -e 'create database my_database' | ||
| + | |||
| + | 4) load the database with the script <span class="pops">[http://gmod.cvs.sourceforge.net/viewvc/*checkout*/gmod/Generic-Genome-Browser/bin/gbrowse_syn/load_alignment_database.pl?pathrev=stable load_alignment_database.pl]</span> | ||
| + | |||
| + | gunzip -c processed_alignments.txt.gz | perl load_alignment_database.pl | ||
Revision as of 12:15, 14 January 2009
Schema
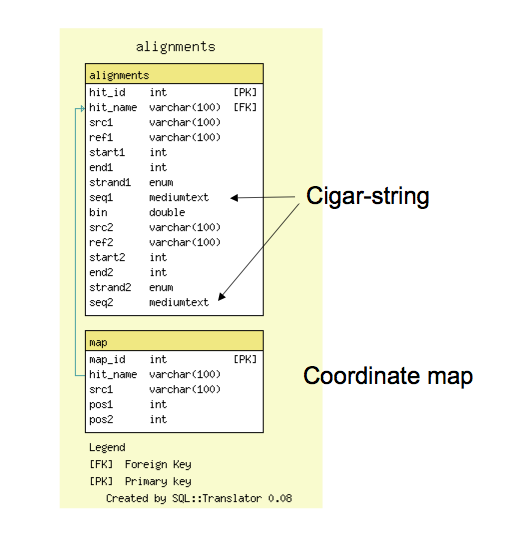
- The alignment database is very simple; it has a tables for all reciprocal 'hits', or alignment features and a table for (optional) 1:1 coordinate maps
- The alignments table contains coordinate information and also support cigar-line representations and the alignment to facilitate future reconstruction of the alignment within GBrowse_syn.
Loading
- The starting point for loading the alignment database is a CLUSTALW format multiple sequence alignment
- The source of the alignment data is up to you but the supported format for entry is currently CLUSTALW
Clustal alignment format
CLUSTAL W(1.81) multiple sequence alignment
c_briggsae-chrII(+)/43862-46313 ATGAGCTTCCACAAAAGCATGAGCTTTCTCAGCTTCTGCCACATCAGCATTCAAATGATC
c_remanei-Crem_Contig172(-)/123228-124941 ATGAGCCTCTACAACCGCATGATTCTTTTCAGCCTCTGCCACGTCCGCATTCAAATGCTC
c_brenneri-Cbre_Contig60(+)/627772-630087 ATGAGCCTCCACAACAGCATGATTTTTCTCGGCTTCCGCCACATCCGCATTCAAATGATC
c_elegans-II(+)/9706834-9708803 ATGAGCCTCTACTACAGCATGATTCTTCTCAGCTTCTGCAACGTCAGCATTCAGATGATC
****** ** ** * ****** ** ** ** ** ** ** ** ******* *** **
c_briggsae-chrII(+)/43862-46313 CGCACAAATATGATGCACAAATCCACAACCTAAAGCATCTCCGATAACGTTGACCGAAGT
c_remanei-Crem_Contig172(-)/123228-124941 AGCACAAATGTAATGAACGAATCCGCATCCCAACGCATCGCCAATCACATTCACAGATGT
c_brenneri-Cbre_Contig60(+)/627772-630087 CGCACAAATGTAGTGGACAAATCCGCATCCCAAAGCGTCTCCGATAACATTTACCGAAGT
c_elegans-II(+)/9706834-9708803 TGCACAAATGTGATGAACGAATCCACATCCCAATGCATCACCGATCACATTGACAGATGT
******** * ** ** ***** ** ** ** ** ** ** ** ** ** ** ** **
c_briggsae-chrII(+)/43862-46313 CCGGAGTCGATCCCTGAAT-----------------------------------------
c_remanei-Crem_Contig172(-)/123228-124941 ACGAAGTCGGTCCCTATAAGGTATGATTTTATATGA----TGTACCATAAGGAAATAGTC
c_brenneri-Cbre_Contig60(+)/627772-630087 ACGAAGTCGATCCCTGAAA---------TCAGATGAGCGGTTGACCA---GAGAACAACC
c_elegans-II(+)/9706834-9708803 ACGAAGTCGGTCCCTGAAC--AATTATTT----TGA----TATA---GAAAGAAACGGTA
** ***** ***** *
NOTE: The sequence naming convention "species-seqid(strand)/start-end" shown in the above example is essential for the data to be loaded correctly
Database Loading Scripts
1) split large clustal files into one alignment/file with the script split_clustal.pl
gunzip my_huge_clustal_file.aln.gz | perl split_clustal.pl /path/to/smaller_alignment_files
2) parse the alignments using the script clustal2hits.pl
perl clustal2hit.pl /path/to/smaller_alignment_files/*.aln |gzip -c >processed_alignments.txt.gz
The output of this script is an ad hoc intermediate format that encodes the alignment coordinates plus 1:1 mapping of coordinates within the alignment (to facilitate accurate grid-lines in GBrowse_syn)
#species1 seqid1 start1 end1 strand1 reserved species2 seqid2 start2 end2 strand2 reserved c_briggsae chrI 1583997 1590364 + . c_remanei Crem_Contig24 631879 634679 - . \ # pos1-1 pos1-2 ... posn-1 posn-2 | # pos1-2 pos1-1 ... posn-2 posn-1 1584000 634676 1584100 634584 (truncated for display...) | 631900 1590333 632000 1590233 (truncated for display...)
3) create a mysql database
mysql -uroot -ppassword -e 'create database my_database'
4) load the database with the script load_alignment_database.pl
gunzip -c processed_alignments.txt.gz | perl load_alignment_database.pl