|
|
| Line 1: |
Line 1: |
| | | | |
| | + | ==Description== |
| | + | |
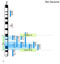
| | + | Flash GViewer is a '''customizable Flash movie''' that can be easily inserted into a web page to display each chromosome in a genome along with the locations of individual features on the chromosomes. It is '''intended to provide an overview of the genomic locations of a specific set of features''' - eg. genes and QTLs associated with a specific phenotype, etc. rather than as a way to view all features on the genome. The features can hyperlink out to a detail page to enable to GViewer to be used as a navigation tool. In addition the bands on the chromosomes can link to defineable URL and new region selection sliders can be used to select a specific chromosome region and then link out to a genome browser for higher resolution information. |
| | | | |
| − | {| width="100%"
| |
| − | |
| |
| − | ; '''[[Comparative Map]]]Description'''<br />
| |
| − | : Flash GViewer is a '''customizable Flash movie''' that can be easily<br /> inserted into a web page to display each chromosome in a genome along<br /> with the locations of individual features on the chromosomes. It is '''intended<br /> to provide an overview of the genomic locations of a specific set of<br /> features''' - eg. genes and QTLs associated with a specific phenotype,<br /> etc. rather than as a way to view all features on the genome. The features<br /> can hyperlink out to a detail page to enable to GViewer to be used<br /> as a navigation tool. In addition the bands on the chromosomes can<br /> link to defineable URL and new region selection sliders can be used<br /> to select a specific chromosome region and then link out to a genome<br /> browser for higher resolution information.
| |
| − | |-
| |
| − | |
| |
| − | '''Current features (v0.4) include:'''
| |
| − | :* Uses a base map containing the chromosomes and banding pattern<br /> of the desired organism. Perl scripts are available to take the UCSC<br /> cytogenetic map files and convert them to the appropriate XML format.<br /> This can also be used to portray any other custom backdrop for a<br /> chromosome - colored segments corresponding to syntenic regions in<br /> other organisms, colored according to feature density, etc.
| |
| − | :* Bands can link out to a URL defined in the base map (to allow users<br /> to click to view a region in a genome browser, for example).
| |
| − | :* Annotations can come from a static XML file or can be dynamically<br /> generated by a server-side script
| |
| − | :* Features appear at the appropriate position on the chromosome and<br /> then dynamically move out to form a non-overlapping display.
| |
| − | :* Placing the mouse over individual features brings up a tooltip<br /> displaying the feature's name, it also visually highlights the region<br /> of the chromosome that the feature is aligned with
| |
| − | :* Clicking on a feature toggles the highlight display so it will<br /> stay on when you move the mouse away. This can be used to turn on<br /> the region highlights for multiple features to better visualize their<br /> overlapping regions. Click again to turn each feature's highlight<br /> off.
| |
| | | | |
| − | * Clicking on a feature can open up a new browser window with a<br /> cusomizable URL, eg a gene report page.
| + | ==Demo & Screenshots== |
| − | * Clicking on a chromosome will expand that chromosome and enlarge<br /> and label the features making them easier to view. Click the chromosome<br /> again to return the whole genome view.
| + | |
| − | * Turn on/off labels for zoom view and genome view (press i button<br /> in bottom left corner). Click redraw to refresh the view to show/hide<br /> labels.
| + | |
| − | * <span class="style11">(New in v0.4) </span>Javascript can be used<br /> to dynamically highlight displayed annotations by using triggering<br /> javascript functions in the web page containing the flash movie.<br /> Javascript can be used to query annotation data from the flash movie<br /> and handle the returned data.
| + | |
| − | * Customizable web interface and perl CGI script provided to allow<br /> users to upload annotations and display them on the GViewer against<br /> the base map of their choice.
| + | |
| − | * <span class="style11">(New in v0.4)</span> Region selection sliders<br /> in Zoom View allow the selection of a genomic region so you can<br /> then click to open up a genome browser window for the specified<br /> region.
| + | |
| | | | |
| − | ;
| + | A live version of the Flash movie is available on the [http://www.gmod.org/nondrupal/FlashGViewer_forWeb/index.html Flash GViewer documentation], screenshots of the tool are provided below. |
| − | :
| + | |
| − | ; '''Demo & Screenshots'''<br />
| + | |
| − | : A live version of the Flash movie is available on the [http://www.gmod.org/nondrupal/FlashGViewer_forWeb/index.html Flash GViewer documentation], screenshots of the tool are provided below.
| + | |
| | |- | | |- |
| | | | | | |
| Line 39: |
Line 21: |
| | | | | | |
| | [[[[Image:gviewer_human_mouse-01.jpg]]]] | | [[[[Image:gviewer_human_mouse-01.jpg]]]] |
| − | |}
| |
| − | |-
| |
| − | |
| |
| − | |-
| |
| − | |
| |
| − | ; '''Requirements'''<br />
| |
| − | : To view the Flash GViewer on a web page requires the Macromedia Flash Plugin version 7, it can be downloaded from Macromedia [http://www.macromedia.com/shockwave/download/download.cgi?P1_Prod_Version=ShockwaveFlash&promoid=BIOW here]. To edit the source code (the .fla file in CVS) and publish modified versions of the .swf requires Macromedia Flash Professional v7.
| |
| − | ; '''Documentation'''<br />
| |
| − | : Installation instructions, explanations of the Flash HTML tags, the XML file formats for the base map and the annotations are listed in the [http://www.gmod.org/nondrupal/FlashGViewer_forWeb/index.html Flash GViewer documentation].<br />
| |
| − | ; '''Contact'''<br />
| |
| − | : [[Simon Twigger]].
| |
| − | ; '''Downloads'''<br />
| |
| − | : [http://sourceforge.net/project/showfiles.php?group_id=27707&package_id=161280 Via the web from sourceforge]
| |
| | |} | | |} |
| | | | |
| − | : <br /><br />
| + | ==Requirements== |
| − | ;
| + | |
| − | :
| + | |
| | | | |
| | + | * Macromedia Flash Plugin version 7, it can be downloaded from Macromedia [http://www.macromedia.com/shockwave/download/download.cgi?P1_Prod_Version=ShockwaveFlash&promoid=BIOW here]. To edit the source code (the .fla file in CVS) and publish modified versions of the .swf requires Macromedia Flash Professional v7. |
| | | | |
| | + | ==Documentation== |
| | | | |
| − | {| id="attachments"
| + | Installation instructions, explanations of the Flash HTML tags, the XML file formats for the base map and the annotations are listed in the [http://www.gmod.org/nondrupal/FlashGViewer_forWeb/index.html Flash GViewer documentation]. |
| − | ! Attachment
| + | |
| − | ! Size
| + | ==Contact== |
| − | |- class="dark"
| + | |
| − | |
| + | [[Simon Twigger]] |
| − | [http://www.gmod.org/files/gviewer_example1-01.jpg gviewer_example1-01.jpg] | + | |
| − | | 1.49 KB
| + | |
| − | |- class="light"
| + | ==Downloads== |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_example1.jpg gviewer_example1.jpg]
| + | [http://sourceforge.net/project/showfiles.php?group_id=27707&package_id=161280 Via the web from sourceforge] |
| − | | 23.04 KB
| + | |
| − | |- class="dark"
| + | |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_example2-01.jpg gviewer_example2-01.jpg]
| + | |
| − | | 1.06 KB
| + | |
| − | |- class="light"
| + | |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_example2.jpg gviewer_example2.jpg]
| + | |
| − | | 11.01 KB
| + | |
| − | |- class="dark"
| + | |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_example_hs-01.jpg gviewer_example_hs-01.jpg]
| + | |
| − | | 1.29 KB
| + | |
| − | |- class="light"
| + | |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_example_hs.jpg gviewer_example_hs.jpg]
| + | |
| − | | 23.46 KB
| + | |
| − | |- class="dark"
| + | |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_human_mouse-01.jpg gviewer_human_mouse-01.jpg] | + | |
| − | | 1.94 KB
| + | |
| − | |- class="light"
| + | |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_human_mouse.jpg gviewer_human_mouse.jpg] | + | |
| − | | 26.23 KB
| + | |
| − | |- class="dark"
| + | |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_rat_human.jpg gviewer_rat_human.jpg]
| + | |
| − | | 26.63 KB
| + | |
| − | |- class="light"
| + | |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_r_m_chr2-01.jpg gviewer_r_m_chr2-01.jpg]
| + | |
| − | | 1.57 KB
| + | |
| − | |- class="dark"
| + | |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_r_m_chr2.jpg gviewer_r_m_chr2.jpg]
| + | |
| − | | 22.27 KB
| + | |
| − | |- class="light"
| + | |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_regionbar.jpg gviewer_regionbar.jpg] | + | |
| − | | 15.67 KB
| + | |
| − | |- class="dark"
| + | |
| − | |
| + | |
| − | [http://www.gmod.org/files/gviewer_regionbar-01.jpg gviewer_regionbar-01.jpg]
| + | |
| − | | 1.33 KB
| + | |
| − | |}
| + | |
Installation instructions, explanations of the Flash HTML tags, the XML file formats for the base map and the annotations are listed in the Flash GViewer documentation.