NOTE: We are working on migrating this site away from MediaWiki, so editing pages will be disabled for now.
WebApollo Installation
Please note that WebApollo has not been officially released, and this guide is development.
Introduction
This guide will walk you through the server side installation for WebApollo. WebApollo is a web-based application, so the only client side requirement is a web browser. Note that WebApollo has only been tested on Chrome, Firefox, and Safari. It currently does not support Internet Explorer.
Installation
All installation steps will be done through a shell. We'll be using Tomcat 7 as our servlet container and PostgreSQL as our relational database management system. We'll use the sample data provided as a separate download.
Server operating system
Any Unix like system (e.g., Unix, Linux, Mac OS X)
Prerequisites
- Servlet container (must support servlet spec 3.0+) [officially supported: Tomcat 7]
- Relational Database Management System [officially supported: PostgreSQL]
- User database helper scripts
- Perl modules
- DBI
- DBD module for your specific RDBMS [officially supported: DBD::Pg for PostgresSQL]
- Perl modules
- Data generation pipeline (see JBrowse prerequisites for more information on its prerequisites)
- Perl modules
- BioPerl 1.6
- JSON
- JSON::XS (optional, for speed)
- PerlIO::gzip
- Heap::Simple
- Heap::Simple::XS
- Devel::Size
- Bio::GFF3::LowLevel::Parser
- System packages
- libpng12-0
- libpng12-dev
- Perl modules
- Sequence search (optional)
- Blat (along with a configured search database)
Conventions
This guide will use the following conventions to make it more concise:
- $WEB_APOLLO_DIR
- Location where the tarball was uncompressed and will include WebApollo-RELEASE_DATE (e.g., ~/webapollo/WebApollo-2012-10-08)
- $WEB_APOLLO_SAMPLE_DIR
- Location where the sample tarball was uncompressed (e.g., ~/webapollo/webapollo_sample)
- $WEB_APOLLO_DATA_DIR
- Location for WebApollo annotations (e.g., /data/webapollo/annotations)
- $JBROWSE_DATA_DIR
- Location for JBrowse data (e.g., /data/webapollo/jbrowse/data)
- $TOMCAT_LIB_DIR
- Location where Tomcat libs are installed (e.g., /usr/share/tomcat7/lib)
- $TOMCAT_CONF_DIR
- Location where Tomcat configuration is installed (e.g., /etc/tomcat7)
- $TOMCAT_WEBAPPS_DIR
- Location where deployed servlets for Tomcat go (e.g., /var/lib/tomcat7/webapps)
- $BLAT_DIR
- Location where the Blat binaries are installed (e.g., /usr/local/bin)
- $BLAT_TMP_DIR
- Location for temporary Blat files (e.g., /data/webapollo/blat/tmp)
- $BLAT_DATABASE
- Location for the Blat database (e.g., /data/webapollo/blat/db/pyu.2bit)
The Tomcat related paths are the ones used by default in Ubuntu 12.04 and Ubuntu's provided Tomcat7 package. Paths will likely be different in your system depending on how Tomcat was installed.
Installation
Uncompress the WebApollo.tgz tarball.
$ tar -xvzf WebApollo-RELEASE_DATE.tgz
User database
WebApollo uses a database to determine who can access and edit annotations for a given sequence.
First we’ll need to create a database. You can call it whatever you want (remember the name as you’ll need to point the configuration to it). For the purposes of this guide, we’ll call it web_apollo_users You might want to create a separate account to manage the database. We’ll have the user web_apollo_user_admin with password web_apollo_user_admin who has database creation privilege. We’ll assume that you’re creating the user database in the same server where WebApollo is being installed (“localhost”).
$ createdb -U web_apollo_users_admin web_apollo_users
Now that the database is created, we need to load the schema to it.
$ cd $WEB_APOLLO_DIR/tools/user $ psql -U web_apollo_users_admin web_apollo_users < user_database_postgresql.sql
Now the user database has been setup.
Let's populate the database.
First we’ll create an user with access to WebApollo. We’ll use the add_user.pl script in $WEB_APOLLO_DIR/tools/user. Let’s create an user named web_apollo_admin with the password web_apollo_admin.
$ ./add_user.pl -D web_apollo_users -U web_apollo_users_admin -P web_apollo_users_admin \
-u web_apollo_admin -p web_apollo_admin
Next we’ll add the annotation tracks ids for the genomic sequences for our organism. We’ll use the add_tracks.pl script in the same directory. We need to generate a file of genomic sequence ids for the script. For convenience, there’s a script called extract_seqids_from_fasta.pl in the same directory which will go through a FASTA file and extract all the ids from the deflines. Let’s first create the list of genomic sequence ids. We'll store it in ~/scratch/seqids.txt. We’ll want to add the prefix “Annotations-” to each identifier.
$ mkdir ~/scratch $ ./extract_seqids_from_fasta.pl -p Annotations- -i ~/$WEB_APOLLO_SAMPLE_DIR/scf1117875582023.fa \ -o ~/scratch/seqids.txt
Now we’ll add those ids to the user database.
$ ./add_tracks.pl -D web_apollo_users -U web_apollo_users_admin -P web_apollo_users_admin \
-t ~/scratch/seqids.txt
Now that we have an user created and the annotation track ids loaded, we’ll need to give the user permissions to access the sequence. We’ll have the all permissions (read, write, publish, user manager). We’ll use the set_track_permissions.pl script in the same directory. We’ll need to provide the script a list of genomic sequence ids, like in the previous step.
$ ./set_track_permissions.pl -D web_apollo_users -U web_apollo_users_admin \
-P web_apollo_users_admin -u web_apollo_admin -t ~/scratch/seqids.txt -a
We’re all done setting up the user database.
Note that we’re only using a subset of the options for all the scripts mentioned above. You can get more detailed information on any given script (and other available options) using the “-h” or “--help” flag when running the script.
Deploying the servlet
Depending on how Tomcat was setup on your server, you might need to run the following command as root.
Note that WebApollo server sends error to the client through JSON messages. Your servlet container must be configured to allow raw JSON to be sent as when errors occur. In the case of Tomcat, you'll need to configure it to use the custom valve that is provided with the WebApollo package.
$ cp $WEB_APOLLO_DIR/tomcat/custom-valves.jar $TOMCAT_LIB_DIR
You'll then need to add errorReportValveClass="org.bbop.apollo.web.ErrorReportValve" as an attribute to the <Host> element in $TOMCAT_CONF_DIR/server.xml
<Host appBase="webapps" autoDeploy="true" name="localhost" unpackWARs="true" errorReportValveClass="org.bbop.apollo.web.ErrorReportValve">We need to deploy the WAR file in the war directory from the unpacked tarball.
$ cd $TOMCAT_WEBAPPS_DIR
Next we need to create the directory that will contain the application.
$ mkdir WebApollo
Now we'll go into the newly created directory and unpack the WAR file into it.
$ cd WebApollo $ jar -xvf $WEB_APOLLO_DIR/war/WebApollo.war
That’s it! We’re done installing WebApollo. Now we need to move on to configuring the application.
Configuration
Most configuration files will reside in $TOMCAT_WEBAPPS_DIR/WebApollo/config. We’ll need to configure a number of things before we can get WebApollo up and running.
Main configuration
The main configuration is stored in $TOMCAT_WEBAPPS_DIR/WebApollo/config/config.xml. Let’s take a look at the file.
<?xml version="1.0" encoding="UTF-8"?> <server_configuration> <!-- mapping configuration for GBOL data structures --> <gbol_mapping>/config/mapping.xml</gbol_mapping> <!-- directory where JE database will be created --> <datastore_directory>ENTER_DATASTORE_DIRECTORY_HERE</datastore_directory> <!-- minimum size for introns created --> <default_minimum_intron_size>1</default_minimum_intron_size> <!-- size of history for each feature - setting to 0 means unlimited history --> <history_size>0</history_size> <!-- overlapping strategy for adding transcripts to genes --> <overlapper_class>org.bbop.apollo.web.overlap.OrfOverlapper</overlapper_class> <!-- class for comparing track names (used for sorting in lists) --> <track_name_comparator_class>org.bbop.apollo.web.track.DefaultTrackNameComparator</track_name_comparator_class> <!-- user authentication/permission configuration --> <user> <!-- database configuration --> <database> <!-- driver for user database --> <driver>org.postgresql.Driver</driver> <!-- JDBC URL for user database --> <url>ENTER_USER_DATABASE_JDBC_URL</url> <!-- username for user database --> <username>ENTER_USER_DATABASE_USERNAME</username> <!-- password for user database --> <password>ENTER_USER_DATABASE_PASSWORD</password> </database> <!-- class for generating user authentication page (login page) --> <authentication_class>org.bbop.apollo.web.user.localdb.LocalDbUserAuthentication</authentication_class> </user> <tracks> <!-- path to JBrowse refSeqs.json file --> <refseqs>ENTER_PATH_TO_REFSEQS_JSON_FILE</refseqs> <!-- annotation track name the current convention is to append the genomic region id to the the name of the annotation track e.g., if the annotation track is called "Annotations" and the genomic region is chr2L, the track name will be "Annotations-chr2L".--> <annotation_track_name>Annotations</annotation_track_name> <!-- organism being annotated --> <organism>ENTER_ORGANISM</organism> <!-- CV term for the genomic sequences - should be in the form of "CV:term". This applies to all sequences --> <sequence_type>ENTER_CVTERM_FOR_SEQUENCE</sequence_type> </tracks> <!-- path to file containing canned comments XML --> <canned_comments>/config/canned_comments.xml</canned_comments> <!-- tools to be used for sequence searching. This is optional. If this is not setup, WebApollo will not have sequence search support --> <sequence_search_tools> <!-- one <sequence_search_tool> element per tool --> <sequence_search_tool> <!-- display name for the search tool --> <key>BLAT nucleotide</key> <!-- class for handling search --> <class>org.bbop.apollo.tools.seq.search.blat.BlatCommandLineNucleotideToNucleotide</class> <!-- configuration for search tool --> <config>/config/blat_config.xml</config> </sequence_search_tool> <sequence_search_tool> <!-- display name for the search tool --> <key>BLAT protein</key> <!-- class for handling search --> <class>org.bbop.apollo.tools.seq.search.blat.BlatCommandLineProteinToNucleotide</class> <!-- configuration for search tool --> <config>/config/blat_config.xml</config> </sequence_search_tool> </sequence_search_tools> </server_configuration>
Let’s look through each element in more detail with values filled in.
<!-- mapping configuration for GBOL data structures --> <gbol_mapping>/config/mapping.xml</gbol_mapping>
File that contains type mappings used by the underlying data model. It’s best not to change the default option.
<!-- directory where JE database will be created --> <datastore_directory>$WEB_APOLLO_DATA_DIR</datastore_directory>
Directory where user generated annotations will be stored. The data is stored using Berkeley DB.
<!-- minimum size for introns created --> <default_minimum_intron_size>1</default_minimum_intron_size>
Minimum length of intron to be created when using the “Make intron” operation. The operation will try to make the shortest intron that’s at least as long as this parameter. So if you set it to a value of “40”, then all calculated introns will be at least of length 40.
<!-- size of history for each feature - setting to 0 means unlimited history --> <history_size>0</history_size>
The size of your history stack, meaning how many “Undo/Redo” steps you can do. The larger the number, the larger the storage space needed. Setting it to “0” makes it to that there’s no limit.
<!-- overlapping strategy for adding transcripts to genes --> <overlapper_class>org.bbop.apollo.web.overlap.OrfOverlapper</overlapper_class>
Defines the strategy to be used for deciding whether overlapping transcripts should be considered splice variants to the same gene. This points to a Java class implementing the org.bbop.apollo.overlap.Overlapper interface. This allows you to create your own custom overlapping strategy should the need arise. Currently available options are:
- org.bbop.apollo.web.overlap.NoOverlapper
- No transcripts should be considered splice variants, regardless of overlap.
- org.bbop.apollo.web.overlap.SimpleOverlapper
- Any overlapping of transcripts will cause them to be part of the same gene
- org.bbop.apollo.web.overlap.OrfOverlapper
- Only transcripts that overlap within the coding region and within frame are considered part of the same gene
<!-- class for comparing track names (used for sorting in lists) --> <track_name_comparator_class>org.bbop.apollo.web.track.DefaultTrackNameComparator</track_name_comparator_class>
Defines how to compare genomic sequence names for sorting purposes in the genomic region selection list. Points to a class implementing the org.bbop.apollo.web.track.TrackNameComparator interface. You can implement your own class to allow whatever sorting you’d like for your own organism. This doesn't make much of a difference in our case since we're only dealing with one genomic region. The only available implementation is:
- org.bbop.apollo.web.track.DefaultTrackNameComparator
- Sorts genomic sequence names lexicographically
Let’s take look at the user element, which handles configuration for user authentication and permission handling.
<!-- user authentication/permission configuration --> <user> <!-- database configuration --> <database> <!-- driver for user database --> <driver>org.postgresql.Driver</driver> <!-- JDBC URL for user database --> <url>ENTER_USER_DATABASE_JDBC_URL</url> <!-- username for user database --> <username>ENTER_USER_DATABASE_USERNAME</username> <!-- password for user database --> <password>ENTER_USER_DATABASE_PASSWORD</password> </database> <!-- class for generating user authentication page (login page) --> <authentication_class>org.bbop.apollo.web.user.localdb.LocalDbUserAuthentication</authentication_class> </user>
Let’s first look at the database element that defines the database that will handle user permissions (which we created previously).
<!-- driver for user database --> <driver>org.postgresql.Driver</driver>
This should point the JDBC driver for communicating with the database. We’re using a PostgreSQL driver since that’s the database we’re using for user permission management.
<!-- JDBC URL for user database --> <url>jdbc:postgresql://localhost/web_apollo_users</url>
JDBC URL to the user permission database. We'll use jdbc:postgresql://localhost/web_apollo_users since the database is running in the same server as the annotation editing engine and we named the database web_apollo_users.
<!-- username for user database --> <username>web_apollo_users_admin</username>
User name that has read/write access to the user database. The user with access to the user database has the user name web_apollo_users_admin.
<!-- password for user database --> <password>web_apollo_users_admin</password>
Password to access user database. The user with access to the user database has the password </tt>web_apollo_users_admin</tt>.
Now let’s look at the other elements in the user element.
<!-- class for generating user authentication page (login page) --> <authentication_class>org.bbop.apollo.web.user.localdb.LocalDbUserAuthentication</authentication_class>
Defines how user authentication is handled. This points to a class implementing the org.bbop.apollo.web.user.UserAuthentication interface. This allows you to implement any type of authentication you’d like (e.g., LDAP). Currently available options are:
- org.bbop.apollo.web.user.localdb.LocalDbUserAuthentication
- Uses the user permission database to also store authentication information, meaning it stores user passwords in the database
- org.bbop.apollo.web.user.browserid.BrowserIdUserAuthentication
- Uses Mozilla’s BrowserID service for authentication. This has the benefits of offloading all authentication security to Mozilla and allows one account to have access to multiple resources (as long as they have BrowserID support). Being that the service is provided through Mozilla, it will require users to create a BrowserID account
Now let’s look at the configuration for accessing the annotation tracks for the genomic sequences.
<tracks> <!-- path to JBrowse refSeqs.json file --> <refseqs>ENTER_PATH_TO_REFSEQS_JSON_FILE</refseqs> <!-- annotation track name the current convention is to append the genomic region id to the the name of the annotation track e.g., if the annotation track is called "Annotations" and the genomic region is chr2L, the track name will be "Annotations-chr2L".--> <annotation_track_name>Annotations</annotation_track_name> <!-- organism being annotated --> <organism>ENTER_ORGANISM</organism> <!-- CV term for the genomic sequences - should be in the form of "CV:term". This applies to all sequences --> <sequence_type>ENTER_CVTERM_FOR_SEQUENCE</sequence_type> </tracks>
Let’s look at each element individually.
<!-- path to JBrowse refSeqs.json file --> <refseqs>$TOMCAT_WEBAPPS_DIR/WebApollo/jbrowse/data/refSeqs.json</refseqs>
Location where the refSeqs.json file resides, which is created from the data generation pipeline (see the data generation section). By default, the JBrowse data needs to reside in $TOMCAT_WEBAPPS_DIR/WebApollo/jbrowse/data. If you want the data to reside elsewhere, you’ll need to do configure your servlet container to handle the appropriate alias to jbrowse/data or symlink the data directory to somewhere else. WebApollo is pre-configured to allow symlinks.
<annotation_track_name>Annotations</annotation_track_name>
Name of the annotation track. Leave it as the default value of Annotations.
<!-- organism being annotated --> <organism>Pythium ultimum</organism>
Scientific name of the organism being annotated (genus and species). We're annotating Pythium ultimum.
<!-- CV term for the genomic sequences - should be in the form of "CV:term". This applies to all sequences --> <sequence_type>sequence:contig</sequence_type>
The type for the genomic sequences. Should be in the form of CV:term. Our genomic sequences are of the type sequence:contig.
<!-- path to file containing canned comments XML --> <canned_comments>/config/canned_comments.xml</canned_comments>
File that contains canned comments (predefined comments that will be available from a pull-down menu when creating comments). It’s best not to change the default option. See the canned comments section for details on configuring canned comments.
<!-- tools to be used for sequence searching. This is optional. If this is not setup, WebApollo will not have sequence search support --> <sequence_search_tools> <!-- one <sequence_search_tool> element per tool --> <sequence_search_tool> <!-- display name for the search tool --> <key>BLAT nucleotide</key> <!-- class for handling search --> <class>org.bbop.apollo.tools.seq.search.blat.BlatCommandLineNucleotideToNucleotide</class> <!-- configuration for search tool --> <config>/config/blat_config.xml</config> </sequence_search_tool> <sequence_search_tool> <!-- display name for the search tool --> <key>BLAT protein</key> <!-- class for handling search --> <class>org.bbop.apollo.tools.seq.search.blat.BlatCommandLineProteinToNucleotide</class> <!-- configuration for search tool --> <config>/config/blat_config.xml</config> </sequence_search_tool> </sequence_search_tools>
Here’s the configuration for sequence search tools (allows searching your genomic sequences). WebApollo does not implement any search algorithms, but instead relies on different tools and resources to handle searching (this provides much more flexible search options). This is optional. If it’s not configured, WebApollo will not have sequence search support. You'll need one sequence_search_tool element per search tool. Let's look at the element in more detail.
<!-- display name for the search tool --> <key>BLAT nucleotide</key>
This is a string that will be used for the display name for the search tool, in the pull down menu that provides search selection for the user.
<!-- class for handling search --> <class>org.bbop.apollo.tools.seq.search.blat.BlatCommandLineNucleotideToNucleotide</class>
Should point to the class that will handle the search request. Searching is handled by classes that implement the org.bbop.apollo.tools.seq.search.SequenceSearchTool interface. This allows you to add support for your own favorite search tools (or resources). We currently only have support for command line Blat, in the following flavors:
- org.bbop.apollo.tools.seq.search.blat.BlatCommandLineNucleotideToNucleotide
- Blat search for a nucleotide query against a nucleotide database
- org.bbop.apollo.tools.seq.search.blat.BlatCommandLineProteinToNucleotide
- Blat search for a protein query against a nucleotide database
<!-- configuration for search tool --> <config>/config/blat_config.xml</config>
File that contains the configuration for the searching plugin chosen. If you’re using Blat, you should not change this. If you’re using your own plugin, you’ll want to point this to the right configuration file (which will be dependent on your plugin). See the Blat section for details on configuring WebApollo to use Blat.
Canned comments
You can configure a set of predefined comments that will be available for users when adding comments through a dropdown menu. The configuration is stored in /usr/local/tomcat/tomcat7/webapps/WebApollo/config/canned_comments.xml. Let’s take a look at the configuration file.
<?xml version="1.0" encoding="UTF-8"?> <canned_comments> <!-- one <comment> element per comment. it must contain the attribute "feature_type" that defines the type of feature this comment will apply to. must be be in the form of "CV:term" (e.g., "sequence:gene") <comment feature_type="sequence:gene">This is a comment for sequence:gene</comment> --> </canned_comments>
You’ll need one <comment> element for each predefined comment. The element needs to have a feature_type attribute in the form of CV:term that this comment applies to. Let’s make a few comments for feature of type sequence:gene and sequence:transcript:
<comment feature_type="sequence:gene">This is a comment for a gene</comment> <comment feature_type="sequence:gene">This is another comment for a gene</comment> <comment feature_type="sequence:transcript">This is a comment for a transcript</comment>
Search tools
As mentioned previously, WebApollo makes use of tools for sequence searching rather than employing its own search algorithm. The only currently supported tool is command line Blat.
Blat
You’ll need to have Blat installed and a search database with your genomic sequences available to make use of this feature. The configuration is stored in $TOMCAT_WEBAPPS_DIR/WebApollo/config/blat_config.xml. Let’s take a look at the configuration file:
<?xml version="1.0" encoding="UTF-8"?> <!-- configuration file for setting up command line Blat support --> <blat_config> <!-- path to Blat binary → <blat_bin>ENTER_PATH_TO_BLAT_BINARY</blat_bin> <!-- path to where to put temporary data --> <tmp_dir>ENTER_PATH_FOR_TEMPORARY_DATA</tmp_dir> <!-- path to Blat database --> <database>ENTER_PATH_TO_BLAT_DATABASE</database> <!-- any Blat options (directly passed to Blat) e.g., -minMatch --> <blat_options>ENTER_ANY_BLAT_OPTIONS</blat_options> </blat_config>
Let’s look at each element with values filled in.
<!-- path to Blat binary --> <blat_bin>$BLAT_DIR/blat</blat_bin>
We need to point to the location where the Blat binary resides. For this guide, we'll assume Blat in installed in /usr/local/bin.
<!-- path to where to put temporary data --> <tmp_dir>$BLAT_TMP_DIR</tmp_dir>
We need to point to the location where to store temporary files to be used in the Blat search. It can be set to whatever location you’d like.
<!-- path to Blat database --> <database>$BLAT_DATABASE</database>
We need to point to the location of the search database to be used by Blat. See the Blat documentation for more information on generation search databases.
<!-- any Blat options (directly passed to Blat) e.g., -minMatch --> <blat_options>-minScore=100 -minIdentity=60</blat_options>
Here we can configure any extra options to used by Blat. These options are passed verbatim to the program. In this example, we’re passing the -minScore parameter with a minimum score of 100 and the -minIdentity parameter with a value of 60 (60% identity). See the Blat documentation for information of all available options.
Data generation
The steps for generating data (in particular static data) are mostly similar to JBrowse data generation steps, with some extra steps required. The scripts for data generation reside in $TOMCAT_WEBAPPS_DIR/WebApollo/jbrowse/bin. Let's go into WebApollo's JBrowse directory.
$ cd $TOMCAT_WEBAPPS_DIR/WebApollo/jbrowse
It will make things easier if we make sure that the scripts in the bin directory are executable.
$ chmod 755 bin/*
As mentioned previously, the data resides in the data directory by default. We can symlink $JBROWSE_DATA_DIR giving you a lot of flexibility in allowing your WebApollo instance to easily point to a new data directory.
$ ln -sf $JBROWSE_DATA_DIR data
Now that we have our data directory in JBrowse, we need to copy some files into it that are specific to WebApollo's JBrowse. We need to copy all of the contents from $WEB_APOLLO_DIR/WebApollo/json in our data directory.
$ cp $WEB_APOLLO_DIR/WebApollo/json/* data
DNA track setup
The first thing we need to do before processing our evidence is to generate the reference sequence data to be used by JBrowse. We'll use the prepare-refseqs.pl script.
$ bin/prepare-refseqs.pl --fasta $WEB_APOLLO_SAMPLE_DIR/scf1117875582023.fa
We now have the DNA track setup.
Static data generation
Generating data from GFF3 works best by having a separate GFF3 per source type. If your GFF3 has all source types in the same file, we need to split up the GFF3. We can use the split_gff_by_source.pl script in $WEB_APOLLO_DIR/tools/data to do so. We'll output the split GFF3 to some temporary directory (we'll use $WEB_APOLLO_SAMPLE_DIR/split_gff).
$ mkdir -p $WEB_APOLLO_SAMPLE_DIR/split_gff $ $WEB_APOLLO_DIR/tools/data/split_gff_by_source.pl -i $WEB_APOLLO_SAMPLE_DIR/scf1117875582023.gff -d $WEB_APOLLO_SAMPLE_DIR/split_gff
If we look at the contents of $WEB_APOLLO_SAMPLE_DIR/split_gff, we can see we have the following files:
$ ls $WEB_APOLLO_SAMPLE_DIR/split_gff blastn.gff est2genome.gff protein2genome.gff repeatrunner.gff blastx.gff maker.gff repeatmasker.gff snap_masked.gff
We need to process each file and create the appropriate tracks.
We'll start off with maker.gff. We need to handle that file a bit differently than the rest of the files since the GFF represents the features as gene, transcript, exons, and CDSs.
$ bin/flatfile-to-json.pl --gff ~/scratch/split_gff/maker.gff \ --arrowheadClass trellis-arrowhead --getSubfeatures \ --subfeatureClasses '{"wholeCDS": "null", "CDS":"maker-CDS", "UTR": "maker-UTR", "exon":"maker-exon"}' \ --cssClass maker-transcript --type mRNA --trackLabel maker
Now we need to convert the Maker track into WebApollo compatible JSON. We can use json_converter.sh in ~/WebApollo/tools/data to do so.
$ ~/WebApollo/tools/data/json_converter.sh -d data/tracks/maker root track dir: data/tracks/maker editing files in place seq dir count: 1 done processing features, total scanned: 353
Lastly we need to modify the maker entry in data/trackList.json. Let's look at the stock entry.
{ "autocomplete" : "none", "style" : { "className" : "maker-transcript", "subfeatureClasses" : { "wholeCDS" : "null", "exon" : "maker-exon", "CDS" : "maker-CDS", "UTR" : "maker-UTR" }, "arrowheadClass" : "trellis-arrowhead" }, "key" : "maker", "phase" : null, "urlTemplate" : "tracks/maker/{refseq}/trackData.json", "compress" : 0, "label" : "maker", "type" : "FeatureTrack", "subfeatures" : 1 }
We need to add "renderClassName" : "maker-transcript-render" to the style member. We also need to change type from FeatureTrack to DraggableFeatureTrack. This is how the updated entry should look like:
{ "autocomplete" : "none", "style" : { "renderClassName" : "maker-transcript-render", "className" : "maker-transcript", "subfeatureClasses" : { "wholeCDS" : "null", "exon" : "maker-exon", "CDS" : "maker-CDS", "UTR" : "maker-UTR" }, "arrowheadClass" : "trellis-arrowhead" }, "key" : "maker", "phase" : null, "urlTemplate" : "tracks/maker/{refseq}/trackData.json", "compress" : 0, "label" : "maker", "type" : "DraggableFeatureTrack", "subfeatures" : 1 }
Now we need to process the other remaining GFF3 files. The entries in those are stored as "match/match_part", so they can all be handled in a similar fashion.
We'll start off with blastn as an example.
$ bin/flatfile-to-json.pl --gff ~/scratch/split_gff/blastn.gff \ --arrowheadClass trellis-arrowhead --getSubfeatures \ --subfeatureClasses '{"match_part": "blastn-alignment-part"}' \ --cssClass "blastn-alignment-match" --trackLabel blastn
Now we need to modify the blastn entry in data/trackList.json. Let's look at the stock entry.
{ "autocomplete" : "none", "style" : { "className" : "blastn-match", "subfeatureClasses" : { "match_part" : "blastn-part" }, "arrowheadClass" : "trellis-arrowhead" }, "key" : "blastn", "phase" : null, "urlTemplate" : "tracks/blastn/{refseq}/trackData.json", "compress" : 0, "label" : "blastn", "type" : "FeatureTrack", "subfeatures" : 1 }
We need to add "renderClassName" : "blastn-alignment-render" to the style member. We also need to change type from FeatureTrack to DraggableFeatureTrack. This is how the updated entry should look like:
{ "autocomplete" : "none", "style" : { "renderClassName" : "blastn-alignment-render", "className" : "blastn-match", "subfeatureClasses" : { "match_part" : "blastn-part" }, "arrowheadClass" : "trellis-arrowhead" }, "key" : "blastn", "phase" : null, "urlTemplate" : "tracks/blastn/{refseq}/trackData.json", "compress" : 0, "label" : "blastn", "type" : "DraggableFeatureTrack", "subfeatures" : 1 }
We need to follow the same steps for the remaining GFF3 files. It can be a bit tedious to do this for the remaining six files, so we can use a simple inline shell script to help us out. However, we'll still have to manually modify data/trackList.json for each new track. Don't worry if the script doesn't make sense, you can still process each file by hand. For now you can just copy and paste the script into your terminal.
$ for i in $(ls ~/scratch/split_gff/*.gff | grep -v maker); do j=$(basename $i); j=${j/.gff/}; echo "Processing $j" && bin/flatfile-to-json.pl --gff $i --arrowheadClass trellis-arrowhead \ --getSubfeatures --subfeatureClasses "{\"match_part\": \"$j-alignment-part\"}" \ --cssClass "$j-alignment-match" --trackLabel $j; done
IMPORTANT!
Remember that we still need to add "renderClassName" : "$type-alignment-render" to the style member, where $type is the track name. We also need to change type from FeatureTrack to DraggableFeatureTrack.
Now let's look how to configure BAM support. WebApollo has native support for BAM, so extra processing of the data is required.
First we'll copy the BAM data into the WebApollo data directory. We'll put it in the data/bam directory. Keep in mind that this BAM data was randomly generated, so there's really no biological meaning to it. We only created it to show BAM support.
$ mkdir data/bam $ cp ~/maker_output/*.bam* data/bam
Now we need to edit the data/bam_trackList.json file. Let's look at the file.
{ "tracks" : [ { "type" : "BamFeatureTrack", "label" : "ENTER_LABEL", "data_url" : "ENTER_URL_TO_BAM_FILE", "index_url" : "ENTER_URL_TO_BAI_FILE", "config" : { "autocomplete" : "none", "style" : { "className" : "bam", "subfeatureClasses" : { "M": "cigarM", "D": "cigarD", "N": "cigarN", "=": "cigarEQ", "E": "cigarEQ", "X": "cigarX", "I": "cigarI" }, "arrowheadClass" : null }, "key" : "ENTER_KEY", "compress" : 0, "type" : 1, "subfeatures" : 1 } } ], "formatVersion" : 1 }
We need to modify the following elements: label, data_url, index_url, and key.
Let's look at each value with their values filled in.
"label" : "simulated_bam",
The internal name for this BAM track.
"data_url" : "
bam/scf1117875582023.bam ",
URL to BAM file.
"index_url" : "
bam/scf1117875582023.bam.bai ",
URL to BAM index file.
"sourceUrl" : "data/",
Add sourceUrl under index_url.
You can point to the BAM/BAI file in any server by providing an absolute URL. This is a nice feature since BAM files can get rather large, so you don't need to create a local copy on your own server.
"key" : "simulated BAM",
The external, human-readable label seen on the BAM track.
Here's how the updated JSON should look like.
{ "tracks" : [ { "type" : "BamFeatureTrack", "label" : "simulated_bam", "data_url" : "bam/scf1117875582023.bam", "index_url" : "bam/scf1117875582023.bam.bai", "sourceUrl" : "data/", "config" : { "autocomplete" : "none", "style" : { "className" : "bam", "subfeatureClasses" : { "M": "cigarM", "D": "cigarD", "N": "cigarN", "=": "cigarEQ", "E": "cigarEQ", "X": "cigarX", "I": "cigarI" }, "arrowheadClass" : null }, "key" : "simulated BAM", "compress" : 0, "type" : 1, "subfeatures" : 1 } } ], "formatVersion" : 1 }
You should now have a simulated BAM track now available.
You can download the updated trackList.json file here (you'll need to put the file in your virtual machine and unzip it in
/usr/local/tomcat/tomcat7/webapps/WebApollo/jbrowse/data):
web_apollo_track_list_json.zip
Alternatively, you can just copy the WebApolloDemo trackList.json into WebApollo (since they're both using the same dataset).
$ cd /usr/local/tomcat/tomcat7/webapps/WebApollo/jbrowse/data $ cp /usr/local/tomcat/tomcat7/webapps/WebApolloDemo/jbrowse/data/trackList.json .
Congratulations, you're done configuring WebApollo.
Let's test out our installation. Point your browser to
http:// ec2-##-##-##-##.compute-1.amazonaws.com:8080/WebApollo
(e.g.,
http://ec2-184-73-92-243.compute-1.amazonaws.com:8080/WebApollo ).
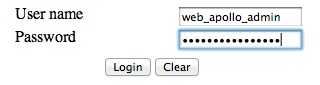
The user name and password are both web_apollo_admin as we configured earlier.
Click on the Edit annotations button.
We only see one reference sequence to annotate since we're only working with one contig. Click on the Edit button.
Now have fun annotating!!!