GMOD
Sybil Chado Comparative Genomics Data
Sybil Chado Comparative Genomics Data storage
This page will detail the TIGR/JCVI/IGS Sybil way of storing comparative genomics data.
The following diagram describes how protein clusters are stored for Sybil:
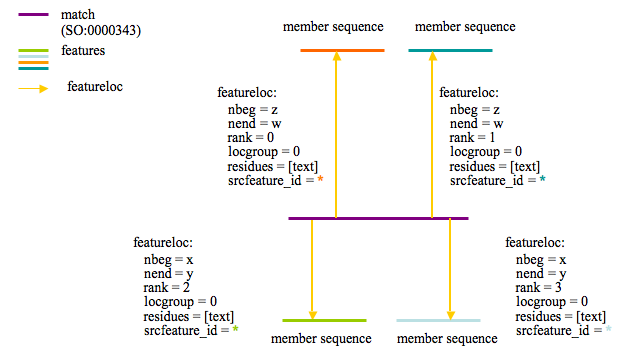
The purple line represents the protein cluster and is stored in feature.
The proteins that are members of the cluster are shown with the labels ‘member sequence’ and are linked to the cluster feature via featurloc (where feature_id = protein feature and srcfeature_id = cluster feature). The featureloc rank column is incremented for each cluster member.
Cluster assignments are based on bi-directional best BLASTP hit. Top BLASTP hits are stored in the database via ‘match’ and ‘match_part’ features independent of the clustering analysis. In this way multiple clustering analyses may be loaded which use the same set of BLASTP hits. Loading multiple clustering analyses can be useful when one wants to try out/compare some new clustering parameters or would like to cluster on only a subset of the loaded genomes.
The use of featureloc to join matching regions allows for the method to include storage of locatable syntenic regions based on whole genome alignment or other genomic DNA based comparison methods. Doing this would simply require replacing the ‘polypeptide’ feature with an ‘assembly’ feature in the featureloc table.
I propose adding another link to this scheme as I feel it could help with performance and simplify some queries. The new relationship would be as follows:
A feature_relationship of type derived_from (Seems the most appropriate?) that links the cluster Match feature with each of the blastp match features that were used to generate this cluster. The new picture would look like this:
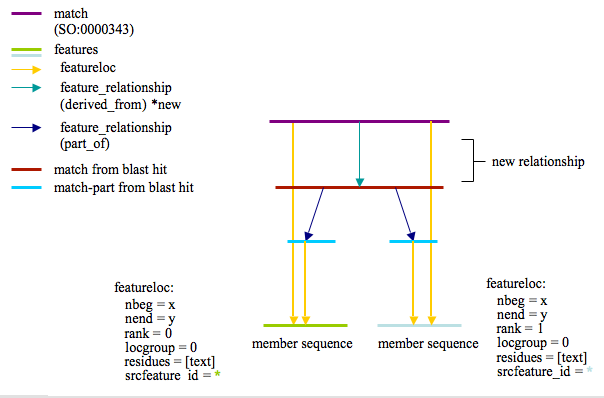
Proposed schema extensions related to Sybil/IGS needs