GMOD
Chado Mage Module
Contents
- 1 Introduction
- 2
Tables
- 2.1 Table: acquisition
- 2.2 Table: acquisition_relationship
- 2.3 Table: acquisitionprop
- 2.4 Table: arraydesign
- 2.5 Table: arraydesignprop
- 2.6 Table: assay
- 2.7 Table: assay_biomaterial
- 2.8 Table: assay_project
- 2.9 Table: assayprop
- 2.10 Table: biomaterial
- 2.11 Table: biomaterial_dbxref
- 2.12 Table: biomaterial_relationship
- 2.13 Table: biomaterial_treatment
- 2.14 Table: biomaterialprop
- 2.15 Table: channel
- 2.16 Table: control
- 2.17 Table: element
- 2.18 Table: element_relationship
- 2.19 Table: elementresult
- 2.20 Table: elementresult_relationship
- 2.21 Table: magedocumentation
- 2.22 Table: mageml
- 2.23 Table: protocol
- 2.24 Table: protocolparam
- 2.25 Table: quantification
- 2.26 Table: quantification_relationship
- 2.27 Table: quantificationprop
- 2.28 Table: study
- 2.29 Table: study_assay
- 2.30 Table: studydesign
- 2.31 Table: studydesignprop
- 2.32 Table: studyfactor
- 2.33 Table: studyfactorvalue
- 2.34 Table: treatment
Introduction
The Mage module is designed to store data from microarray experiments. It is based on the RAD database but has been substantially modified to contain the necessary foreign keys and satisfy the Chado naming conventions. The Mage module is compatible with the MAGE standard.
Other Documentation
See the Mage FAQ.
Mage and Expression
The Mage module and the Expression module can be considered overlapping but complementary. The Mage module can store data taken directly from the experimental results whereas the Expression module is typically used to store summary data taken from the biological literature, or extracted from the microarray data stored in Mage. The Mage module handles details about experiments that the Expression module does not whereas the Expression module can be thought of a simpler set of tables designed to tie ontologies concerned with expression to sequence features.
Entering and Querying
A typical case would be that the researcher had run some number of microarray experiments. She will need to load the resultant data into the Mage module and related tables, then query them. These tasks could be accomplished using some application but for illustrative purposes we will interact directly with the schema. Let’s assume that the tissues being assayed come from Drosophila melanogaster. The relevant tables are shown below, the purpose is to show the relationships between the key tables.
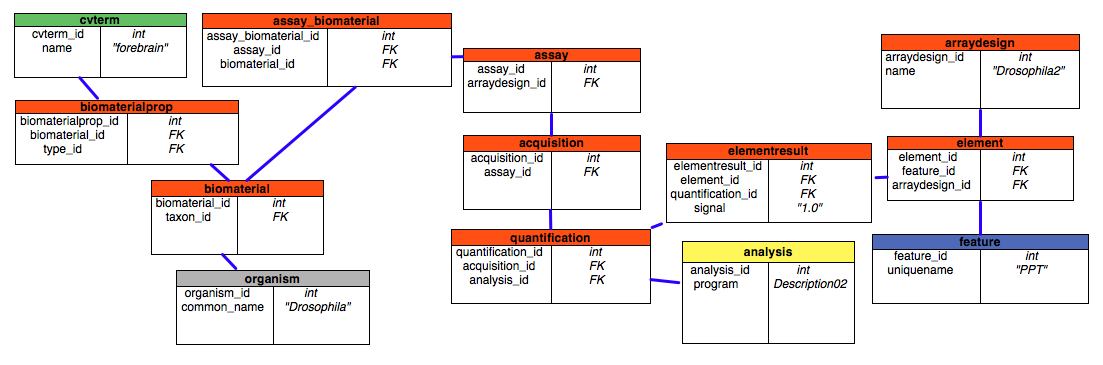
Red is for the mage tables, Yellow is for companalysis tables, Gray is for organism tables, Blue is for sequence tables, and Green is for the cv tables.
Loading
The researcher would load the data into the database in this order:
- Assume that forebrain, or any other relevant ontology terms, are records in the cvterm table from a previously loaded anatomy ontology.
- Create a biomaterial record for the forebrain sample the expression was observed in. The organism_id would be for Drosophila melanogaster (assume all the relevant species are already loaded in organism).
- Create a biomaterialprop record to link records from 1 (cvterm) and 2 ( biomaterial).
- Create or use an arraydesign record for the assay platform. This could be something like Drosophila2 (an Affymetrix platform), or even a string like features if we just want to report expression or lack thereof for all genes in the assayed sample.
- Create an assay record to represent the event where the forebrain sample was measured. It links to the record created in 4 (arraydesign).
- Link records from 2 (biomaterial) and 5 (assay) in assay_biomaterial. The relationship here is many-to-many between assays and biomaterials because of multichannel and multiplexed assay technology.
- Create an acquisition record that depends on 5 (assay). This is how the assay’s results were digitized, typically using a digital camera or scanner, but it can refer to any data acquired from the assay in general.
- Create an analysis record. This is the algorithm that is used to process the data from 7 (acquisition).
- Create a quantification record. It depends on 7 (acquisition) and 8 (analysis), and represents data from 7 processed using 8.
- Create element records, one per gene that is assayable using 4 (arraydesign). Each element record has a nullable attribute where it can point back to feature records to associate elements directly with genomic features.
- Create elementresult records, one for each record created in 10 (element) and pointing back to 9 (quantification) which ultimately links back to the sample. Experimental result data is stored here.
You can store a boolean for ‘expressed’ or ‘not expressed’, or you could store the quantitative data and have some algorithm that determines from those data what is or is not expressed. Obviously the latter is less lossy but is also less straightforward for the casual observer to interpret.
Querying
Once data has been loaded according to the general approach above the schema can be queryed. A typical question the researcher may ask is “which genes are expressed in the Drosophila forebrain”. That question is roughly equivalent to this SQL query:
SELECT uniquename FROM feature
JOIN element ON feature.feature_id = element.feature_id
JOIN elementresult ON element.element_id = elementresult.element_id
JOIN quantification ON elementresult.quantification_id = quantification.quantification_id
JOIN acquisition ON quantification.acquisition_id = acquisition.acquisition_id
JOIN assay ON acquisition.assay_id = assay.assay_id
JOIN assay_biomaterial ON assay.assay_id = assay_biomaterial.assay_id
JOIN biomaterial ON assay_biomaterial.biomaterial_id = biomaterial.biomaterial_id
JOIN organism ON biomaterial.taxon_id = organism.organism_id
JOIN biomaterialprop ON biomaterial.biomaterial_id = biomaterialprop.biomaterial_id
JOIN cvterm ON biomaterialprop.type_id = cvterm.cvterm_id
WHERE organism.common_name = 'Drosophila' AND
WHERE cvterm.name = 'forebrain' AND
WHERE elementresult.signal > 0;
Tables
Table: acquisition
This represents the scanning of hybridized material. The output of this process is typically a digital image of an array.
| F-Key | Name | Type | Description |
|---|---|---|---|
| acquisition_id | serial | PRIMARY KEY | |
| assay | assay_id | integer | NOT NULL |
| protocol | protocol_id | integer | |
| channel | channel_id | integer | |
| acquisitiondate | timestamp without time zone | DEFAULT (‘now’::text)::timestamp(6) with time zone | |
| name | text | UNIQUE | |
| uri | text |
acquisition Structure
Tables referencing this one via Foreign Key Constraints:
Table: acquisition_relationship
Multiple monochrome images may be merged to form a multi-color image. Red-green images of 2-channel hybridizations are an example of this.
| F-Key | Name | Type | Description |
|---|---|---|---|
| acquisition_relationship_id | serial | PRIMARY KEY | |
| acquisition | subject_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| acquisition | object_id | integer | UNIQUE#1 NOT NULL |
| value | text | ||
| rank | integer | UNIQUE#1 NOT NULL |
acquisition_relationship Structure
Table: acquisitionprop
Parameters associated with image acquisition.
| F-Key | Name | Type | Description |
|---|---|---|---|
| acquisitionprop_id | serial | PRIMARY KEY | |
| acquisition | acquisition_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| value | text | ||
| rank | integer | UNIQUE#1 NOT NULL |
acquisitionprop Structure
Table: arraydesign
General properties about an array. An array is a template used to generate physical slides, etc. It contains layout information, as well as global array properties, such as material (glass, nylon) and spot dimensions (in rows/columns).
| F-Key | Name | Type | Description |
|---|---|---|---|
| arraydesign_id | serial | PRIMARY KEY | |
| contact | manufacturer_id | integer | NOT NULL |
| cvterm | platformtype_id | integer | NOT NULL |
| cvterm | substratetype_id | integer | |
| protocol | protocol_id | integer | |
| dbxref | dbxref_id | integer | |
| name | text | UNIQUE NOT NULL | |
| version | text | ||
| description | text | ||
| array_dimensions | text | ||
| element_dimensions | text | ||
| num_of_elements | integer | ||
| num_array_columns | integer | ||
| num_array_rows | integer | ||
| num_grid_columns | integer | ||
| num_grid_rows | integer | ||
| num_sub_columns | integer | ||
| num_sub_rows | integer |
arraydesign Structure
Tables referencing this one via Foreign Key Constraints:
Table: arraydesignprop
Extra array design properties that are not accounted for in arraydesign.
| F-Key | Name | Type | Description |
|---|---|---|---|
| arraydesignprop_id | serial | PRIMARY KEY | |
| arraydesign | arraydesign_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| value | text | ||
| rank | integer | UNIQUE#1 NOT NULL |
arraydesignprop Structure
Table: assay
An assay consists of a physical instance of an array, combined with the conditions used to create the array (protocols, technician information). The assay can be thought of as a hybridization.
| F-Key | Name | Type | Description |
|---|---|---|---|
| assay_id | serial | PRIMARY KEY | |
| arraydesign | arraydesign_id | integer | NOT NULL |
| protocol | protocol_id | integer | |
| assaydate | timestamp without time zone | DEFAULT (‘now’::text)::timestamp(6) with time zone | |
| arrayidentifier | text | ||
| arraybatchidentifier | text | ||
| contact | operator_id | integer | NOT NULL |
| dbxref | dbxref_id | integer | |
| name | text | UNIQUE | |
| description | text |
assay Structure
Tables referencing this one via Foreign Key Constraints:
Table: assay_biomaterial
A biomaterial can be hybridized many times (technical replicates), or combined with other biomaterials in a single hybridization (for two-channel arrays).
| F-Key | Name | Type | Description |
|---|---|---|---|
| assay_biomaterial_id | serial | PRIMARY KEY | |
| assay | assay_id | integer | UNIQUE#1 NOT NULL |
| biomaterial | biomaterial_id | integer | UNIQUE#1 NOT NULL |
| channel | channel_id | integer | UNIQUE#1 |
| rank | integer | UNIQUE#1 NOT NULL |
assay_biomaterial Structure
Table: assay_project
Link assays to projects.
| F-Key | Name | Type | Description |
|---|---|---|---|
| assay_project_id | serial | PRIMARY KEY | |
| assay | assay_id | integer | UNIQUE#1 NOT NULL |
| project | project_id | integer | UNIQUE#1 NOT NULL |
assay_project Structure
Table: assayprop
Extra assay properties that are not accounted for in assay.
| F-Key | Name | Type | Description |
|---|---|---|---|
| assayprop_id | serial | PRIMARY KEY | |
| assay | assay_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| value | text | ||
| rank | integer | UNIQUE#1 NOT NULL |
assayprop Structure
Table: biomaterial
A biomaterial represents the MAGE concept of BioSource, BioSample, and LabeledExtract. It is essentially some biological material (tissue, cells, serum) that may have been processed. Processed biomaterials should be traceable back to raw biomaterials via the biomaterialrelationship table.
| F-Key | Name | Type | Description |
|---|---|---|---|
| biomaterial_id | serial | PRIMARY KEY | |
| organism | taxon_id | integer | |
| contact | biosourceprovider_id | integer | |
| dbxref | dbxref_id | integer | |
| name | text | UNIQUE | |
| description | text |
biomaterial Structure
Tables referencing this one via Foreign Key Constraints:
Table: biomaterial_dbxref
| F-Key | Name | Type | Description |
|---|---|---|---|
| biomaterial_dbxref_id | serial | PRIMARY KEY | |
| biomaterial | biomaterial_id | integer | UNIQUE#1 NOT NULL |
| dbxref | dbxref_id | integer | UNIQUE#1 NOT NULL |
biomaterial_dbxref Structure
Table: biomaterial_relationship
Relate biomaterials to one another. This is a way to track a series of treatments or material splits/merges, for instance.
| F-Key | Name | Type | Description |
|---|---|---|---|
| biomaterial_relationship_id | serial | PRIMARY KEY | |
| biomaterial | subject_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| biomaterial | object_id | integer | UNIQUE#1 NOT NULL |
biomaterial_relationship Structure
Table: biomaterial_treatment
Link biomaterials to treatments. Treatments have an order of operations (rank), and associated measurements (unittype_id, value).
| F-Key | Name | Type | Description |
|---|---|---|---|
| biomaterial_treatment_id | serial | PRIMARY KEY | |
| biomaterial | biomaterial_id | integer | UNIQUE#1 NOT NULL |
| treatment | treatment_id | integer | UNIQUE#1 NOT NULL |
| cvterm | unittype_id | integer | |
| value | real | ||
| rank | integer | NOT NULL |
biomaterial_treatment Structure
Table: biomaterialprop
Extra biomaterial properties that are not accounted for in biomaterial.
| F-Key | Name | Type | Description |
|---|---|---|---|
| biomaterialprop_id | serial | PRIMARY KEY | |
| biomaterial | biomaterial_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| value | text | ||
| rank | integer | UNIQUE#1 NOT NULL |
biomaterialprop Structure
Table: channel
Different array platforms can record signals from one or more channels (cDNA arrays typically use two CCD, but Affymetrix uses only one).
| F-Key | Name | Type | Description |
|---|---|---|---|
| channel_id | serial | PRIMARY KEY | |
| name | text | UNIQUE NOT NULL | |
| definition | text | NOT NULL |
channel Structure
Tables referencing this one via Foreign Key Constraints:
Table: control
| F-Key | Name | Type | Description |
|---|---|---|---|
| control_id | serial | PRIMARY KEY | |
| cvterm | type_id | integer | NOT NULL |
| assay | assay_id | integer | NOT NULL |
| tableinfo | tableinfo_id | integer | NOT NULL |
| row_id | integer | NOT NULL | |
| name | text | ||
| value | text | ||
| rank | integer | NOT NULL |
control Structure
Table: element
Represents a feature of the array. This is typically a region of the array coated or bound to DNA.
| F-Key | Name | Type | Description |
|---|---|---|---|
| element_id | serial | PRIMARY KEY | |
| feature | feature_id | integer | UNIQUE#1 |
| arraydesign | arraydesign_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | |
| dbxref | dbxref_id | integer |
element Structure
Tables referencing this one via Foreign Key Constraints:
Table: element_relationship
Sometimes we want to combine measurements from multiple elements to get a composite value. Affymetrix combines many probes to form a probeset measurement, for instance.
| F-Key | Name | Type | Description |
|---|---|---|---|
| element_relationship_id | serial | PRIMARY KEY | |
| element | subject_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| element | object_id | integer | UNIQUE#1 NOT NULL |
| value | text | ||
| rank | integer | UNIQUE#1 NOT NULL |
element_relationship Structure
Table: elementresult
An element on an array produces a measurement when hybridized to a biomaterial (traceable through quantification_id). This is the base data from which tables that actually contain data inherit.
| F-Key | Name | Type | Description |
|---|---|---|---|
| elementresult_id | serial | PRIMARY KEY | |
| element | element_id | integer | UNIQUE#1 NOT NULL |
| quantification | quantification_id | integer | UNIQUE#1 NOT NULL |
| signal | double precision | NOT NULL |
elementresult Structure
Tables referencing this one via Foreign Key Constraints:
Table: elementresult_relationship
Sometimes we want to combine measurements from multiple elements to get a composite value. Affymetrix combines many probes to form a probeset measurement, for instance.
| F-Key | Name | Type | Description |
|---|---|---|---|
| elementresult_relationship_id | serial | PRIMARY KEY | |
| elementresult | subject_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| elementresult | object_id | integer | UNIQUE#1 NOT NULL |
| value | text | ||
| rank | integer | UNIQUE#1 NOT NULL |
elementresult_relationship Structure
Table: magedocumentation
| F-Key | Name | Type | Description |
|---|---|---|---|
| magedocumentation_id | serial | PRIMARY KEY | |
| mageml | mageml_id | integer | NOT NULL |
| tableinfo | tableinfo_id | integer | NOT NULL |
| row_id | integer | NOT NULL | |
| mageidentifier | text | NOT NULL |
magedocumentation Structure
Table: mageml
This table is for storing extra bits of MAGEml in a denormalized form. More normalization would require many more tables.
| F-Key | Name | Type | Description |
|---|---|---|---|
| mageml_id | serial | PRIMARY KEY | |
| mage_package | text | NOT NULL | |
| mage_ml | text | NOT NULL |
mageml Structure
Tables referencing this one via Foreign Key Constraints:
Table: protocol
Procedural notes on how data was prepared and processed.
| F-Key | Name | Type | Description |
|---|---|---|---|
| protocol_id | serial | PRIMARY KEY | |
| cvterm | type_id | integer | NOT NULL |
| pub | pub_id | integer | |
| dbxref | dbxref_id | integer | |
| name | text | UNIQUE NOT NULL | |
| uri | text | ||
| protocoldescription | text | ||
| hardwaredescription | text | ||
| softwaredescription | text |
protocol Structure
Tables referencing this one via Foreign Key Constraints:
Table: protocolparam
Parameters related to a protocol. For example, if the protocol is a soak, this might include attributes of bath temperature and duration.
| F-Key | Name | Type | Description |
|---|---|---|---|
| protocolparam_id | serial | PRIMARY KEY | |
| protocol | protocol_id | integer | NOT NULL |
| name | text | NOT NULL | |
| cvterm | datatype_id | integer | |
| cvterm | unittype_id | integer | |
| value | text | ||
| rank | integer | NOT NULL |
protocolparam Structure
Table: quantification
Quantification is the transformation of an image acquisition to numeric data. This typically involves statistical procedures.
| F-Key | Name | Type | Description |
|---|---|---|---|
| quantification_id | serial | PRIMARY KEY | |
| acquisition | acquisition_id | integer | NOT NULL |
| contact | operator_id | integer | |
| protocol | protocol_id | integer | |
| analysis | analysis_id | integer | UNIQUE#1 NOT NULL |
| quantificationdate | timestamp without time zone | DEFAULT (‘now’::text)::timestamp(6) with time zone | |
| name | text | UNIQUE#1 | |
| uri | text |
quantification Structure
Tables referencing this one via Foreign Key Constraints:
Table: quantification_relationship
There may be multiple rounds of quantification, this allows us to keep an audit trail of what values went where.
| F-Key | Name | Type | Description |
|---|---|---|---|
| quantification_relationship_id | serial | PRIMARY KEY | |
| quantification | subject_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| quantification | object_id | integer | UNIQUE#1 NOT NULL |
quantification_relationship Structure
Table: quantificationprop
Extra quantification properties that are not accounted for in quantification.
| F-Key | Name | Type | Description |
|---|---|---|---|
| quantificationprop_id | serial | PRIMARY KEY | |
| quantification | quantification_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| value | text | ||
| rank | integer | UNIQUE#1 NOT NULL |
quantificationprop Structure
Table: study
| F-Key | Name | Type | Description |
|---|---|---|---|
| study_id | serial | PRIMARY KEY | |
| contact | contact_id | integer | NOT NULL |
| pub | pub_id | integer | |
| dbxref | dbxref_id | integer | |
| name | text | UNIQUE NOT NULL | |
| description | text |
study Structure
Tables referencing this one via Foreign Key Constraints:
Table: study_assay
| F-Key | Name | Type | Description |
|---|---|---|---|
| study_assay_id | serial | PRIMARY KEY | |
| study | study_id | integer | UNIQUE#1 NOT NULL |
| assay | assay_id | integer | UNIQUE#1 NOT NULL |
study_assay Structure
Table: studydesign
| F-Key | Name | Type | Description |
|---|---|---|---|
| studydesign_id | serial | PRIMARY KEY | |
| study | study_id | integer | NOT NULL |
| description | text |
studydesign Structure
Tables referencing this one via Foreign Key Constraints:
Table: studydesignprop
| F-Key | Name | Type | Description |
|---|---|---|---|
| studydesignprop_id | serial | PRIMARY KEY | |
| studydesign | studydesign_id | integer | UNIQUE#1 NOT NULL |
| cvterm | type_id | integer | UNIQUE#1 NOT NULL |
| value | text | ||
| rank | integer | UNIQUE#1 NOT NULL |
studydesignprop Structure
Table: studyfactor
| F-Key | Name | Type | Description |
|---|---|---|---|
| studyfactor_id | serial | PRIMARY KEY | |
| studydesign | studydesign_id | integer | NOT NULL |
| cvterm | type_id | integer | |
| name | text | NOT NULL | |
| description | text |
studyfactor Structure
Tables referencing this one via Foreign Key Constraints:
Table: studyfactorvalue
| F-Key | Name | Type | Description |
|---|---|---|---|
| studyfactorvalue_id | serial | PRIMARY KEY | |
| studyfactor | studyfactor_id | integer | NOT NULL |
| assay | assay_id | integer | NOT NULL |
| factorvalue | text | ||
| name | text | ||
| rank | integer | NOT NULL |
studyfactorvalue Structure
Table: treatment
A biomaterial may undergo multiple treatments. Examples of treatments: apoxia, fluorophore and biotin labeling.
| F-Key | Name | Type | Description |
|---|---|---|---|
| treatment_id | serial | PRIMARY KEY | |
| rank | integer | NOT NULL | |
| biomaterial | biomaterial_id | integer | NOT NULL |
| cvterm | type_id | integer | NOT NULL |
| protocol | protocol_id | integer | |
| name | text |
treatment Structure
Tables referencing this one via Foreign Key Constraints: