Difference between revisions of "JBrowse 2 Tutorial PAG 2022"
| Line 1: | Line 1: | ||
This is very much a draft version of the PAG 2022 tutorial, using the JBrowse 1 tutorial as a template. | This is very much a draft version of the PAG 2022 tutorial, using the JBrowse 1 tutorial as a template. | ||
| − | |||
== Prerequisites == | == Prerequisites == | ||
Revision as of 00:53, 29 December 2021
This is very much a draft version of the PAG 2022 tutorial, using the JBrowse 1 tutorial as a template.
Contents
Prerequisites
JBrowse 2 is both a desktop and server application. In this tutorial, we will focus on the desktop application to make our lives easier, but the server application is pretty easy to set up and has simple prerequisites (but reminder: you don't need this for this tutorial):
- a web server like Apache or Nginx
- NodeJS version 10 or better
That's really it for the server. Other things the would likely help include GenomeTools for sorting GFF, SamTools for working with BAM and CRAM files, and tabix for indexing various file.
But, again, none of those things are needed today!
Download and install
While we've installed JBrowse 2 on the conference computers, if you'd like to follow along on your own computer, you can go to https://jbrowse.org/jb2/download/ and get the download for your platform and install it. It shouldn't take very long.
JBrowse Introduction
How and why JBrowse 2 is different from most other web-based genome browsers, including JBrowse and GBrowse.
Replace with current presentation!
Setting up JBrowse
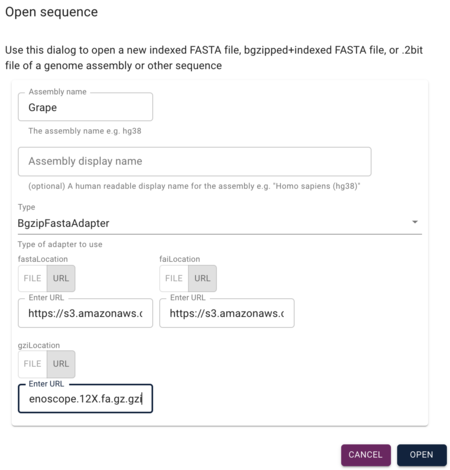
After installing JBrowse 2, open it using your operating systems preferred method, and you'll be greeted with a splash screen that has on part of it, this dialog to open a new sequence:
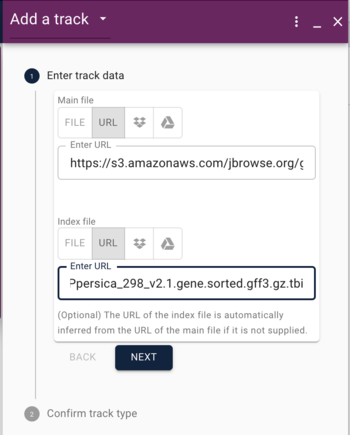
JBrowse supports a variety of forms of sequence data including "vanilla" FASTA, but for this example, we are going to use gzipped and faidx (FASTA indexed) files. To load those up, we'll use the grape FASTA file and it's indexes (ie, 'fai' and 'gzip' files):
https://s3.amazonaws.com/jbrowse.org/genomes/grape/Vvinifera_145_Genoscope.12X.fa.gz https://s3.amazonaws.com/jbrowse.org/genomes/grape/Vvinifera_145_Genoscope.12X.fa.gz.fai https://s3.amazonaws.com/jbrowse.org/genomes/grape/Vvinifera_145_Genoscope.12X.fa.gz.gzi
In the Open Sequence dialog, give the assembly a name (something creative, like "grape") and select BgzipFastaAdapter from the "type" menu, and then copy and paste the above URLs into the appropriate textfields under the "type" menu.
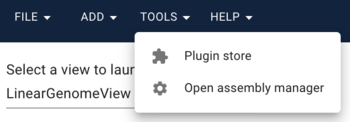
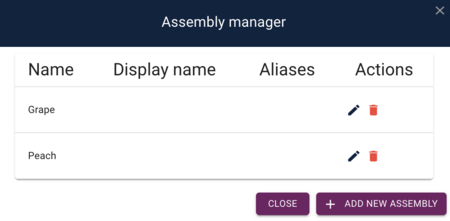
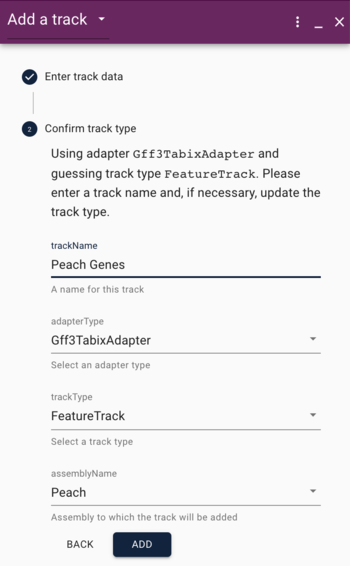
If we were creating a "normal" genome browser, we'd be done with adding sequence, but since we'd like to compare, we will also add the bgzipped and indexed FASTA file for peach. When we clicked on the "open sequence" button before, we were presented with a menu asking us what type of view we'd like, but first we have to add a second genome. What we need is in the Tools menu. Select "Open assembly manager," where you'll get a dialog that was very similar to what we used for grape. This time, we'll load the peach genome, so do the same things as before, and use these URLs:
https://s3.amazonaws.com/jbrowse.org/genomes/peach/Ppersica_298_v2.0.fa.gz https://s3.amazonaws.com/jbrowse.org/genomes/peach/Ppersica_298_v2.0.fa.gz.fai https://s3.amazonaws.com/jbrowse.org/genomes/peach/Ppersica_298_v2.0.fa.gz.gzi
After adding the peach genome, we'll get a dialog that shows us that we have both genomes:
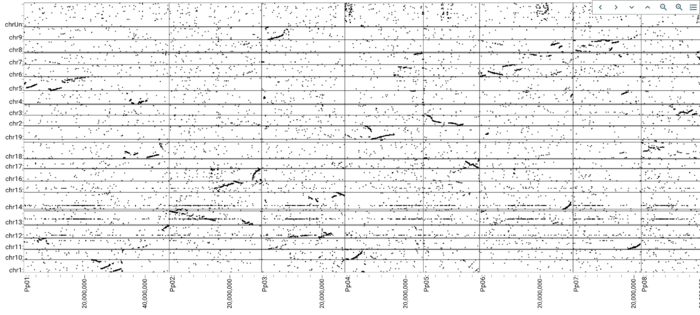
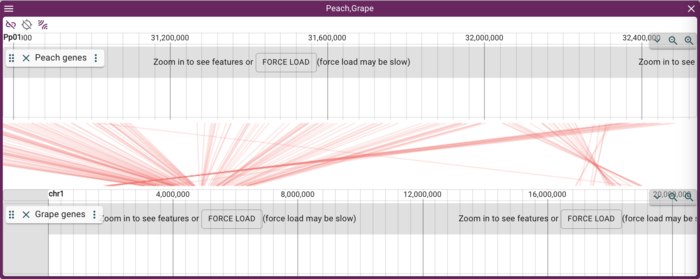
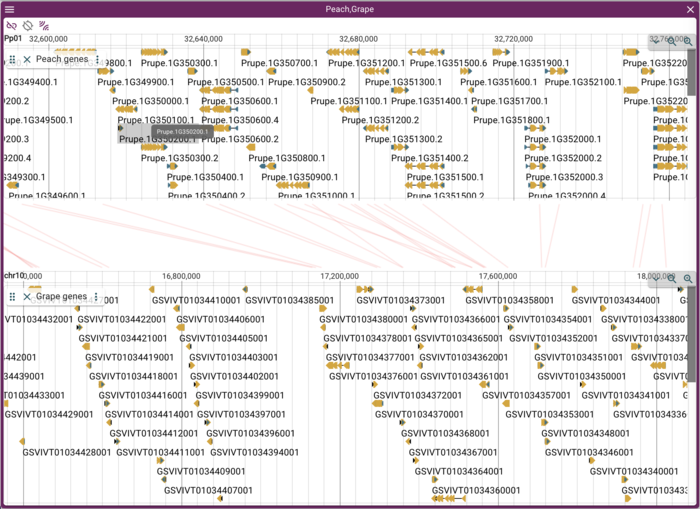
Now we'd like to create a comparison view. JBrowse 2 supports a view comparative views, but we'll start with a whole genome dotplot. For showing areas of synteny, we have a PAF file that looks like this:
Pp01 47851208 1388059 1391133 + chr8 22385789 1539799 1542834 703 3099 21 tp:A:P cm:i:73 s1:i:686 s2:i:439 dv:f:0.1377 rl:i:921840 Pp01 47851208 19134590 19135964 - chr15 20304914 6572992 6574378 659 1387 1 tp:A:P cm:i:85 s1:i:657 s2:i:638 dv:f:0.0768 rl:i:921840 Pp01 47851208 19134614 19135805 + chr17 17126926 16801080 16802270 638 1192 0 tp:A:S cm:i:79 s1:i:638 dv:f:0.0727 rl:i:921840 Pp01 47851208 43719774 43728648 - chr18 29360087 6242566 6251482 642 8964 54 tp:A:P cm:i:55 s1:i:620 s2:i:40 dv:f:0.2275 rl:i:921840 Pp01 47851208 40987755 40994103 + chr18 29360087 2664522 2670983 639 6461 51 tp:A:P cm:i:64 s1:i:620 s2:i:77 dv:f:0.1931 rl:i:921840 Pp01 47851208 19134590 19135968 - chr5 25021643 19591018 19592393 572 1379 0 tp:A:S cm:i:69 s1:i:572 dv:f:0.0910 rl:i:921840
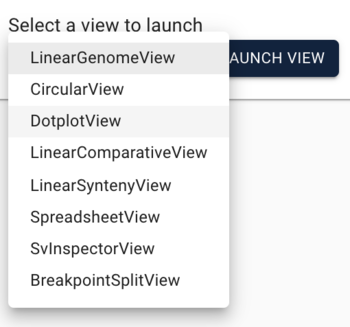
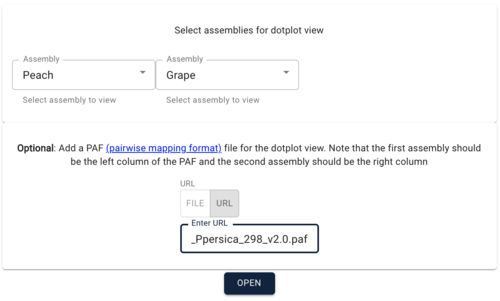
PAF (URL) is a fairly simple file format relating two areas in genome coordinates. To load the peach-grape PAF, select "DotplotView" from the "Select a view to launch" menu.
In the resulting dialog box, select Peach and then Grape for the assemblies to view. IMPORTANT: order here matters! Because the PAF file has the peach coordinates first, you have to use it first in this dialog box. After selecting the two assemblies, copy and paste this URL for the PAF file in to the optional PAF URL textfield:
https://s3.amazonaws.com/jbrowse.org/genomes/synteny/peach_grape.paf
Getting JBrowse
A Short Detour for GFF
GFF (Generic Feature Format) is a very commonly used text format for describing features that exist on sequences. We'll head off to that page to talk about it a bit.