Difference between revisions of "JBrowse 2 Tutorial PAG 2022"
(→Setting up JBrowse) |
(→Setting up JBrowse) |
||
| Line 38: | Line 38: | ||
== Setting up JBrowse == | == Setting up JBrowse == | ||
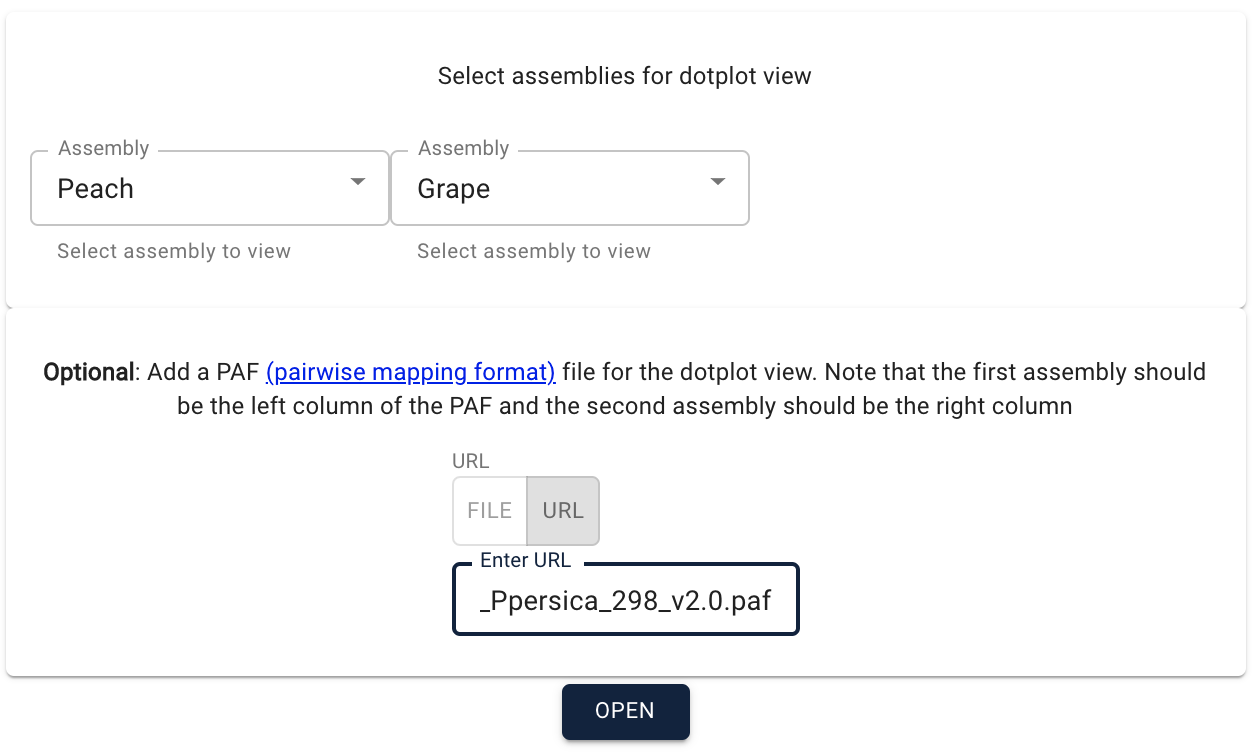
| − | [[File:launch new session.png||Launch new session dialog]] | + | [[File:launch new session.png|350px|Launch new session dialog]] |
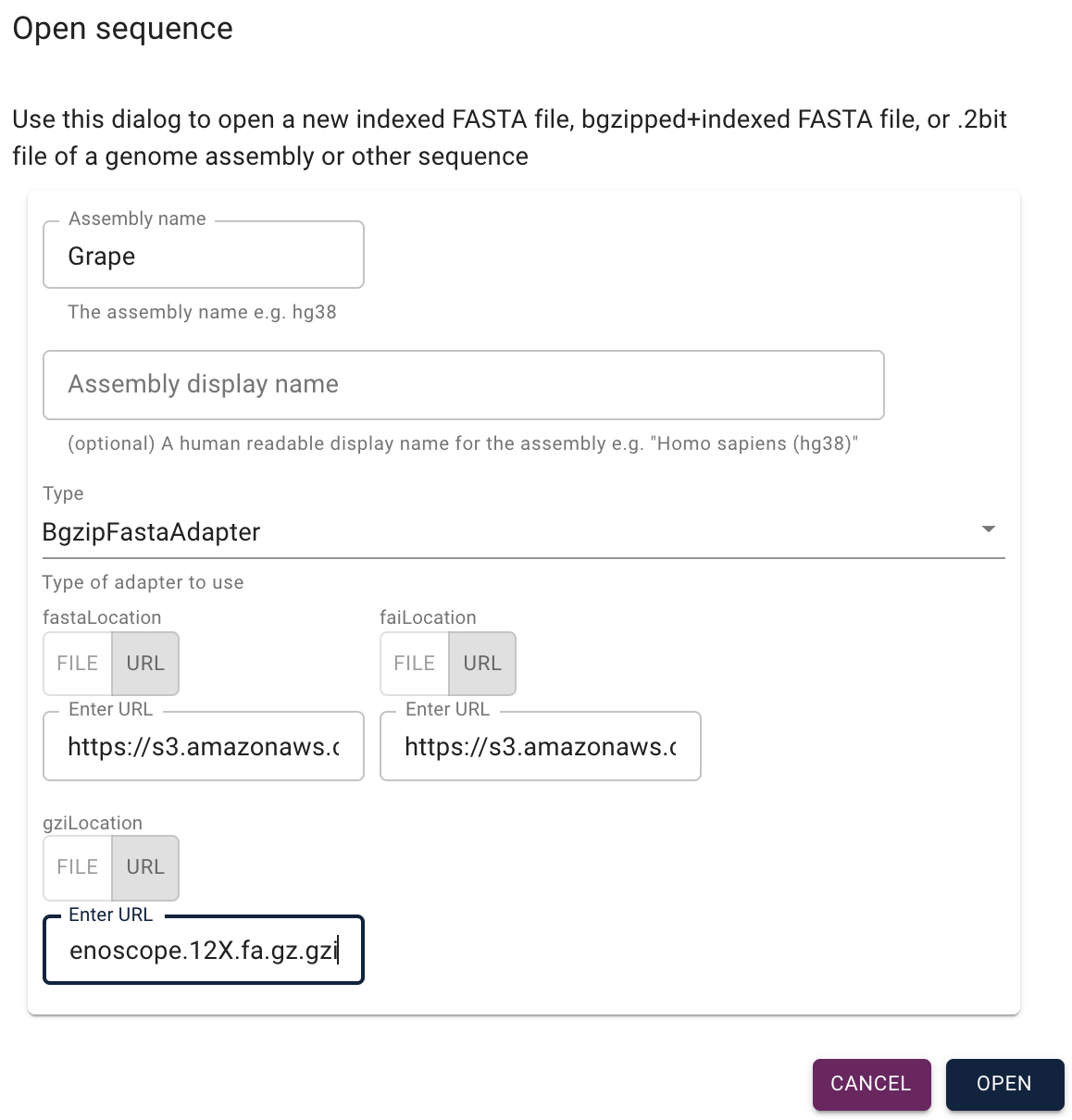
[[File:open new sequence.png||Open a new sequence dialog]] | [[File:open new sequence.png||Open a new sequence dialog]] | ||
Revision as of 22:59, 28 December 2021
This is very much a draft version of the PAG 2022 tutorial, using the JBrowse 1 tutorial as a template.
This tutorial assumes a VirtualBox Ubuntu 18.04 (LTS) instance with the tutorial bundle zip file, also available on Amazon S3: JBrowse PAG 2020.ova (about 4GB) or PAG_2020_JBrowse.zip (about 36MB) for just the JBrowse source and data files for this tutorial.
Contents
Prerequisites
Prerequisites are installed by JBrowse automatically. A few things may fail to install (like legacy support for wiggle files), but that doesn't matter.
Make sure you can copy/paste from the wiki.
It's also very useful to know how to tab-complete in the shell.
It's probably a good idea to use a browser like Chrome or Firefox that has the ability to turn off caching while working on this tutorial. To do this in Chrome, with the browser open to the JBrowse page you're working on, select Developer->Javascript Console from the View menu. In the console, click the "gear" icon (settings) and check the box labeled "Disable Cache".
A few basic packages were installed before JBrowse via apt-get:
sudo apt-get install build-essential zlib1g-dev apache2 curl
Also, a few items were installed that aren't needed for this tutorial but would be necessary if you wanted to add plugins (git and NodeJS):
curl -sL https://deb.nodesource.com/setup_13.x | sudo -E bash - sudo apt-get install -y nodejs sudo apt-get install git
JBrowse Introduction
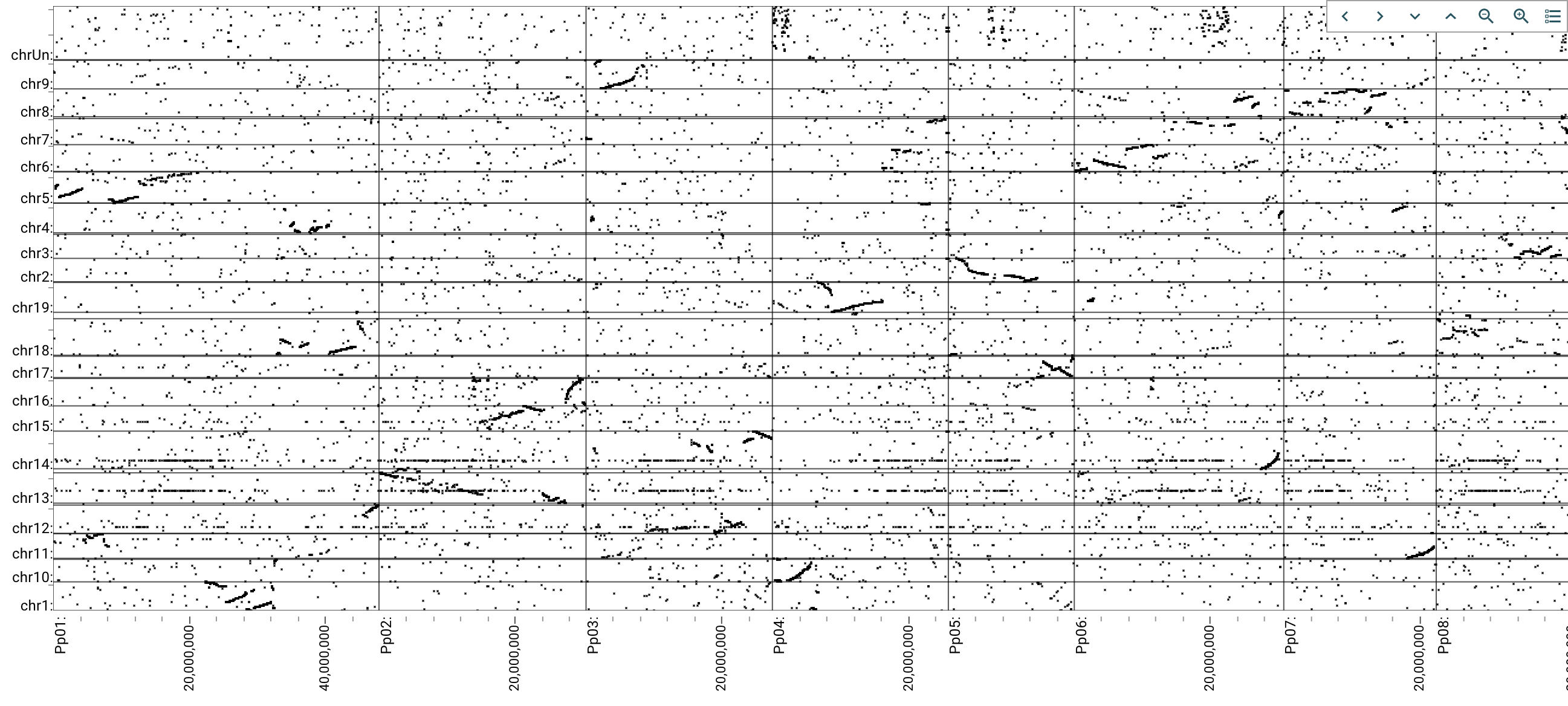
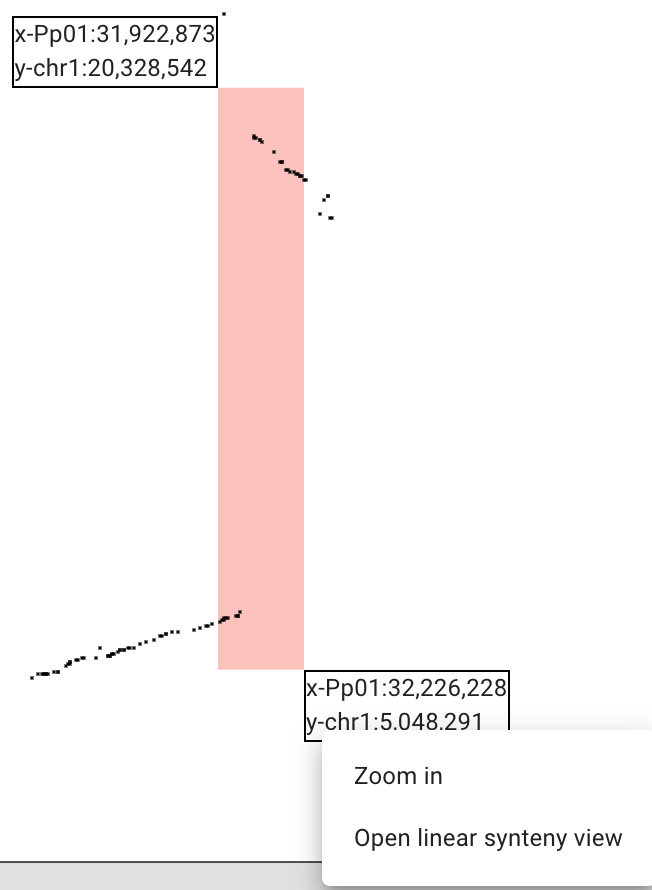
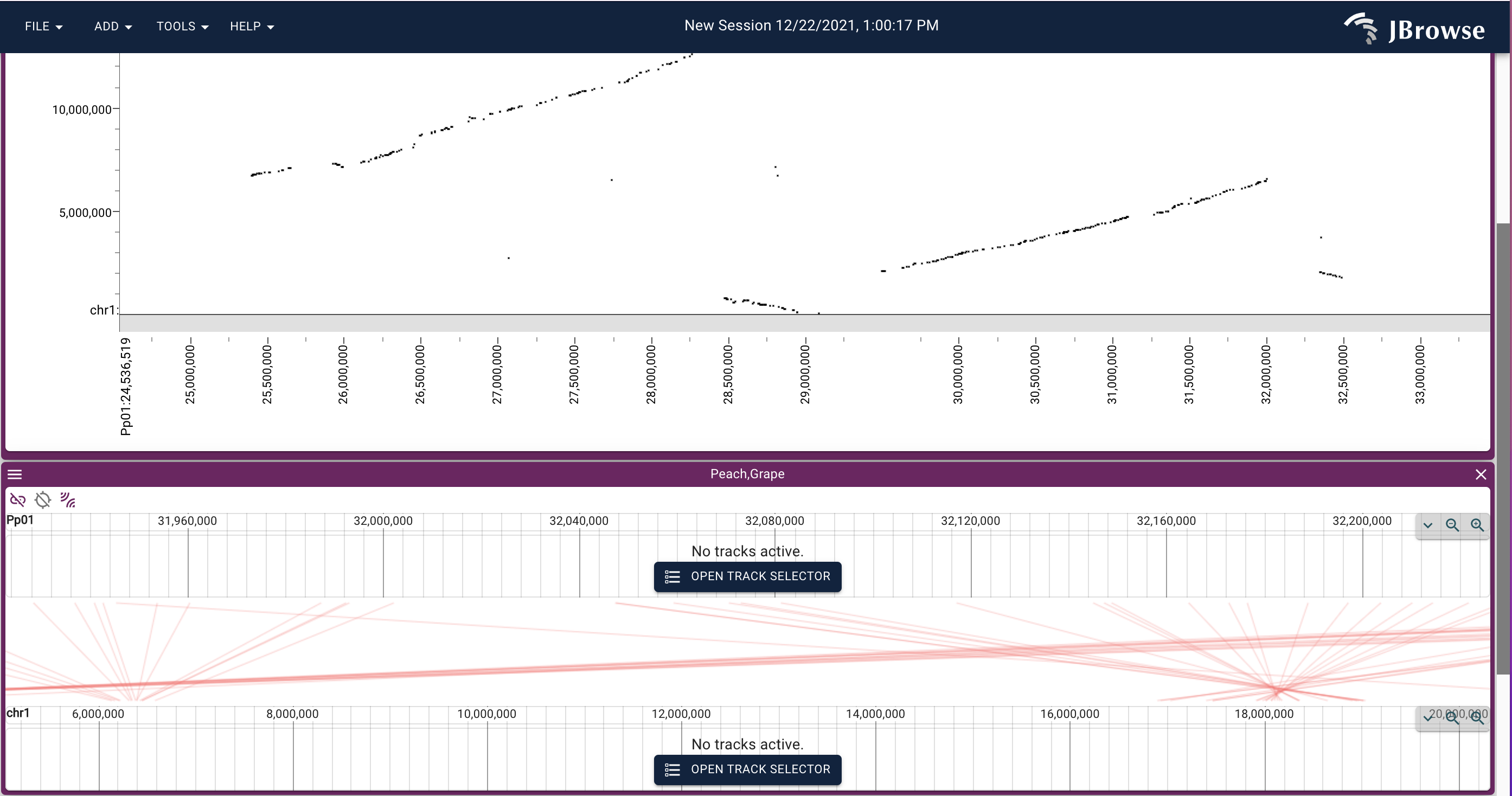
How and why JBrowse is different from most other web-based genome browsers, including GBrowse.
More detail: paper
Setting up JBrowse
Getting JBrowse
A Short Detour for GFF
GFF (Generic Feature Format) is a very commonly used text format for describing features that exist on sequences. We'll head off to that page to talk about it a bit.
Features from a directory of files
Here, we'll use the Bio::DB::SeqFeature::Store adaptor in "memory" mode to read a directory of files. There are adaptors available for use with many other databases, such as Chado and Bio::DB::GFF.
Config file: pythium-1.conf
{
"description": "PAG 2017 P. ultima Example",
"db_adaptor": "Bio::DB::SeqFeature::Store",
"db_args" : {
"-adaptor" : "memory",
"-dir" : ".."
},
...