GBrowse/Uploading Wiggle Tracks
Contents
Dense Feature and Quantitative Data
Wiggle tracks
- Dense feature data includes tiling arrays and other classes of microarray experiments, pre-computed density calculations, or any kind of data that comes in numerous small parts spaced along the full length of the chromosome.
- This wiggle track, originally developed by Jim Kent as part of the wiggle track upload process at the UCSC Genome Browser and has recently been added to Gbrowse.
- Detailed information on the wiggle format can be obtained at the UCSC web site
Note: For the GBrowse wiggle loader to work correctly, you must have the following modules installed:
- Statistics::Descriptive
Format Descriptions
WIG files are plain text files. They always begin with a "track" header, which, at a minimum, looks like this:
track type=wiggle_0 name="ArrayExpt1" description="20 degrees, 2 hr"
The "type" attribute is required, and must have a value of "wiggle_0". "name" and "description" are optional, but suggested, and indicate the name and description of the data series -- these will become the "Name" and "Note" fields of the generated GFF3 feature. Following the track line comes the data for one or more chromosomal regions. As described in the UCSC documentation, there are three ways of formatting the data: (1)"Bed Format", (2) "variableStep", and (3) "fixedStep" format. The BED format is essentially the same as GFF3 and does not give you any performance advantages over using straight GFF3. However, you are dealing with features of variable width and variable steps, BED will allow you to express this. variableStep format describes intervals of the genome that have a fixed width, but begin at arbitrary locations, while fixedStep format describes features of the genome that are evenly spaced and have a fixed width (e.g. tiling array features).
Wiggle (BED)
For BED, the format is:
I 455 480 0 I 495 520 59 I 522 547 0 I 546 571 110 I 576 601 159 I 691 716 189 I 808 833 168 I 834 859 40 I 860 885 0 I 910 935 0 I 932 957 74 I 961 986 0 I 1036 1061 0 I 1067 1092 0 I 1095 1120 88
- There are four required fields: ref_sequence,start,end and score.
- Note that this format differs from the stand-alone BED specification: there are only four fields and the fourth field will only accept numeric data (signed integers, floats, etc)
- Scores can be any sort of numeric data, including integers, negative numbers and floating point.
- Note that the BED coordinates are "zero-indexed", half open, which means the start coordinate is zero-indexed and the end coordinate is 1-indexed. For display or conversion to variableStep wiggle (below), the start coordinate should in incremented up by 1.
- For the intial upload to the website, there is more of a performance hit associated with BED formatted data. After loading, access and rendered performace is equal for all three formats.
- An example of a type of data that would best be represented with BED would be a microarray with varying probe lengths at varying intervals along the chromosome coordinates.
Wiggle (variable step)
For variableStep data, the format is:
variableStep chrom=chr19 span=150
59304701 10.0
59304901 12.5
59305401 15.0
59305601 17.5
59305901 20.0
59306081 17.5
59306301 15.0
59306691 12.5
59307871 10.0
The data is introduced by a line beginning with the keyword "variableStep", and the arguments "chrom" and "span", which indicate the chromosome on which the features are located, and the width of each feature, in base pairs. This is followed by a series of two-element lines indicating the start position of each feature, and its quantitative value. Values can be any sort of numeric data, including integers, negative numbers and floating point.
- Using this format instead of BED will boost performance for data upload.
- An example of data that can best be represented in variable step wiggle format is an oligonucleotide microarray with fixed probe lengths, whose positions are not at fixed intervals.
Wiggle (fixed step)
For fixedStep data, the format is:
fixedStep chrom=chr19 start=59307401 step=300 span=200
1000
900
800
700
600
500
400
300
200
100
The data is introduced by a line beginning with the keyword "fixedStep", and the arguments "chrom", "span", "start" and "step". The first two arguments are the same as before, while "start" and "step" indicate the starting position of the first feature, and the spacing between each feature. This is followed by a numeric value for each step. In this case, we have described 10 features beginning at position 59307401. Each feature begins 300 bp from the next and is 200 bp wide. In practice, this means that the first 200 bp of each interval is filled with known data, while information on the last 100 bp is "missing."
- This is the most constrained format, in terms of coordinates, but it the most efficient for initial upload and processing.
- An example of a type of data that could use fixed step wiggle format is computed bins of fixed width.
Configuring Data Processing and Display
- Some aspects of the data display can be controlled in the wiggle file
Wiggle Track Options Supported by GBrowse
Parameters for wiggle track definition lines All options are placed in a single line separated by spaces:
track type=wiggle_0 name=track_label description=center_label \
visibility=display_mode color=r,g,b altColor=r,g,b \
maxHeightPixels=max:default:min viewLimits=lower:upper \
windowingFunction=max|mean|min|median smoothingWindow=2-16
The track type with version is REQUIRED, and it currently must be wiggle_0:
type wiggle_0
The remaining values are OPTIONAL:
Supported UCSC-style options
name description visibility full|pack # default is full; full = xy plot, pack = density plot color RRR,GGG,BBB # default is 0,0,0 (black) altColor RRR,GGG,BBB # default is 0,0,0 (black) maxHeightPixels max:default:min # default none; Gbrowse uses default, ignores min and max viewLimits lower:upper # default is range found in data windowingFunction maximum|mean|minimum # default is none smoothingWindow 2-16 # default is none; values are in pixels
GBrowse extended data processing options
- Depending on the score distribution and variance, some care must be taken to avoid a loss in display resolution.
- This applies in particular to score distributions with extreme outliers. The reason for this is that the low and high outliers are taken into account when the data are scaled for compression, so small differences (relative the the score range) between scores can become flattened.
- To remedy loss of display resolution, two options are provided.
transform logsquared|logtransform # default is none trim stdev|stdev2|stdevn # default is none
transform: Specify a transform to be performed on all numeric data within this track prior to loading into the binary wig file. This will bring the scores into a normal distribution and improve data scaling and display resolution. Currently, the following two declarations are recognized:
transform=logsquared y’ = log(y**2) for y != 0
y’ = 0 for y == 0
transform=logtransform y' = log(y) for y >= 0
y' = -log(-y) for y < 0
transform=none y’ = y (no transform - the default)
trim: Specify a trimming function to be performed on the data prior to scaling. Trimming will remove outlier scores, which can have the effect of emphasizing differences between mid-range scores. Currently, the following trim functions are recognized:
trim=stdev1 trim to plus/minus 1 standard deviation of the mean
trim=stdev2 trim to plus/minus 2 standard deviations of the mean
...
trim=stdevN trim to plus/minus N standard deviations of the mean; N is an integer
trim=none no trimming (the default)
Formatting Examples
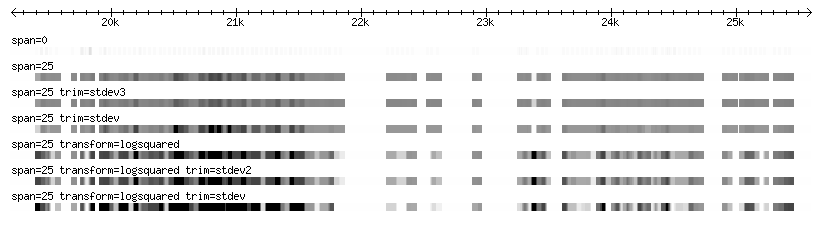
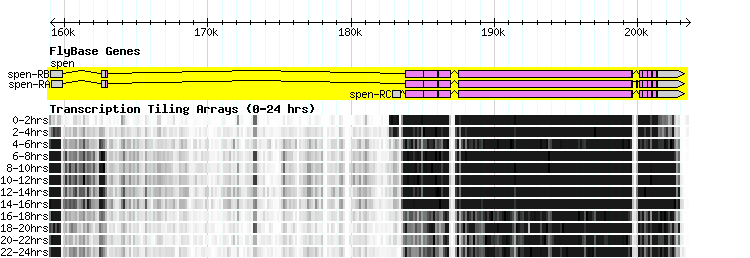
The two images below represent various diplay and loading configurations at high and low magnification for the same set of sample data (Affymetrix tiling data for fly).
Note: The example data used here have an extremely wide range in scores, resulting in a loss of contrast in the majority of the data, which is in the mid-range. Using log transformation and min and max trimming, it is possible to bring out the contrast that is otherwise lost in the middle range.
Zoomed in
- The punctate pattern of the track with no span set is due to each array element being represented as 1bp in width
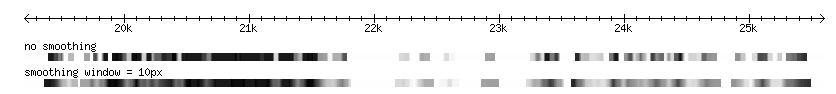
Zoomed out
- Note that the track with no span set is almost entirely lost at this resolution
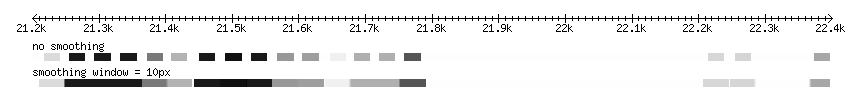
Smoothing
The quantitative data may have a blockish appearance at low magnification. This effect can be minimized by smoothing the data, by blending the transitions between scores within a set number of pixels. The examples below demonstrate smoothing at a resolution of 10px:
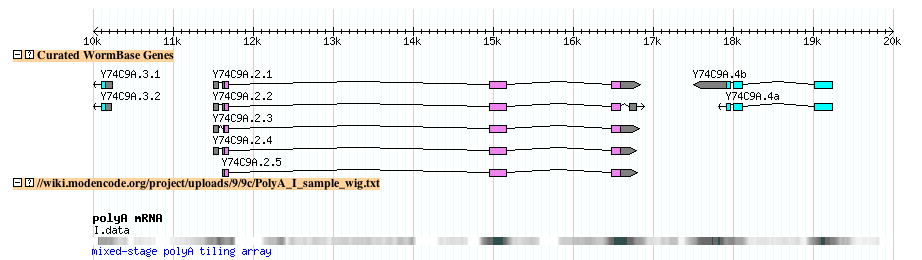
Quantitative Data Examples: C. elegans Tiling Arrays
C. elegans BED
- This is an example of C. elegans tiling array data in BED format (Click here to view the file).
- The first few lines of the example file
track type=wiggle_0 name="polyA" description="mixed-stage polyA tiling array" \ visibility=pack altColor=0,0,255 windowingFunction=mean smoothingWindow=16 I 456 480 0 I 496 520 59 I 523 547 0 I 547 571 110 I 577 601 159 I 692 716 189 I 809 833 168 I 835 859 40 I 861 885 0 I 911 935 0
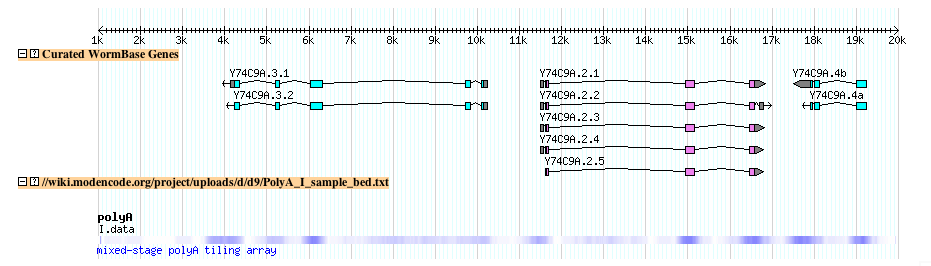
- The file looks lihe this when uploaded to the modENCODE genome browser
- Try it yourself: Follow this link to upload the above example file to the modENCODE C. elegans genome browser
C. elegans Wiggle
- This is an example of C. elegans tiling array data in variable step wiggle format (Click here to view the file).
- The first few lines of the example file:
# Note: comments beginning with '#' are ignored track type=wiggle_0 name="polyA mRNA" description="mixed-stage polyA tiling array"\ color=255,255,255 altColor=0,0,0 windowingFunction=mean smoothingWindow=16 variableStep chrom=I span=25 480 0 520 59 547 0 571 110 601 159 716 189 833 168 859 40 885 0
- This is what the track looks like after uploading to the modENCODE genome browser
Uploading the tracks
Uploading wiggle tracks to GBrowse is accomplished the same way as with other classes of remote data.
In the image below, there are three ways to upload your data:
- Upload a file from your computer via the "Browse..." and "Upload Buttons"
- Paste in your data via the "New..." button
- Put your file up on a web or ftp server and then put the URL (complete with the 'http://' or 'ftp://' protocol directive) in the entry form.
Making a Wiggle Track Permanent
The upload mechanism creates a temporary private track. If you are a GBrowse administrator and wish to create a permanent wiggle track, the process is simple.
- Format and save a wiggle file to disk in the manner described earlier.
- Run the script wiggle2gff3.pl on this file to create the binary wig file. Use the --path option to specify the directory in which you want the binary wig file to be stored (default is the temporary directory), and the --method option to set the feature type (the default is "example"). This will create a binary .wig file in the indicated directory and send a GFF3 file to standard output. You should capture this output using the ">" redirect.
- Load the GFF3 file into your gbrowse database using bp_seqfeature_load.pl or bp_load_gff.pl.
- Configure a stanza for the data using the "wiggle_density" or "wiggle_xyplot" glyphs:
[TEST WIG] feature = example glyph = wiggle_density key = my first wiggle file
If you later need to move the wig file somewhere else, simply edit the GFF3 file to change the path specified in the wigfile attribute. One neat trick is to use a relative path for the wigfile attribute, as in:
wigfile=track003.chr19.1199828298.wig
You can then use the basedir track option to tell the glyph which directory contains the wigfile:
[TEST WIG] feature = example glyph = wiggle_density basedir = /var/data/wigfiles/ key = my first wiggle file
Specifying the Order of Glyphs within a Displayed Track
In the figure at the top of this page, there is a single GBrowse track composed of several horizontal charts, one for each time period. One way to ensure that these charts are displayed in the appropriate order is to use the "source" field in the GFF3 file, in conjunction with the sort order attribute in the stanza for the track.
For example, if the time periods are t=0, t=1, ..., t=9, and the primary source is "Quelle", then the source for the data at t=0 could be called Quelle_0, and similarly for the other time periods, and one would add the following line to the relevant stanza:
sort order = name
Illustrative Script
Assuming there is a .BED file named study_TT.BED for each time period TT, where TT is 00, 01, 02, ...., then the following script illustrates how to generate the .gff3 files for subsequent uploading:
#!/bin/sh
SOURCE=Quelle # a string representing the primary source
STUDY=study # ${STUDY}_$t.BED
METHOD=example # the "feature"
for t in 00 01 02 03 04 05 06 07 08 09 10
do
wiggle2gff3.pl --method $METHOD --source ${SOURCE}_$t ${STUDY}_$t.BED \
> ${STUDY}_$t.gff3
done
Email Threads
- wiggle_xyplot smoothing, 2010/06