NOTE: We are working on migrating this site away from MediaWiki, so editing pages will be disabled for now.
Difference between revisions of "JBrowse 2 Tutorial PAG 2022"
(→JBrowse Introduction) |
(→Setting up JBrowse) |
||
| Line 22: | Line 22: | ||
== Setting up JBrowse == | == Setting up JBrowse == | ||
| + | |||
| + | |||
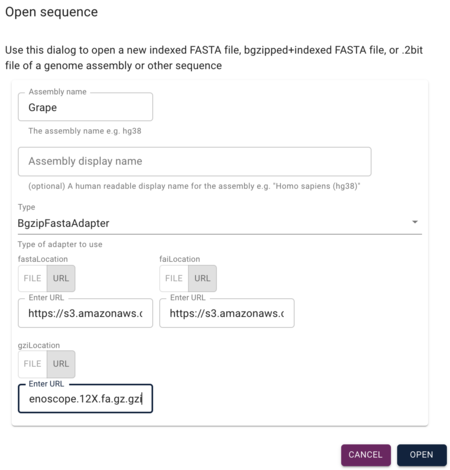
[[File:launch new session.png|border|350px|Launch new session dialog]] | [[File:launch new session.png|border|350px|Launch new session dialog]] | ||
Revision as of 23:45, 28 December 2021
This is very much a draft version of the PAG 2022 tutorial, using the JBrowse 1 tutorial as a template.
This tutorial assumes a VirtualBox Ubuntu 18.04 (LTS) instance with the tutorial bundle zip file, also available on Amazon S3: JBrowse PAG 2020.ova (about 4GB) or PAG_2020_JBrowse.zip (about 36MB) for just the JBrowse source and data files for this tutorial.
Contents
Prerequisites
JBrowse 2 is both a desktop and server application. In this tutorial, we will focus on the desktop application to make our lives easier, but the server application is pretty easy to set up and has simple prerequisites (but reminder, you don't need this for this tutorial):
- a web server like Apache or Nginx
- NodeJS version 10 or better
That's really it for the server. Other things the would likely help include GenomeTools for sorting GFF, SamTools for working with BAM and CRAM files, and tabix for indexing various file.
But, again, none of those things are needed today!
JBrowse Introduction
How and why JBrowse 2 is different from most other web-based genome browsers, including JBrowse and GBrowse.
Replace with current presentation!
Setting up JBrowse
Getting JBrowse
A Short Detour for GFF
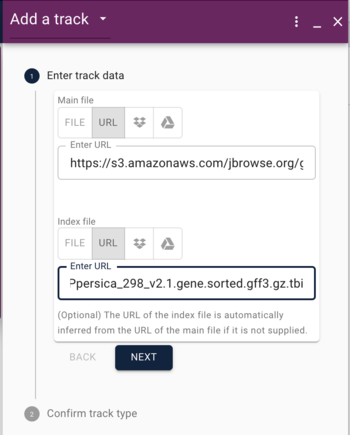
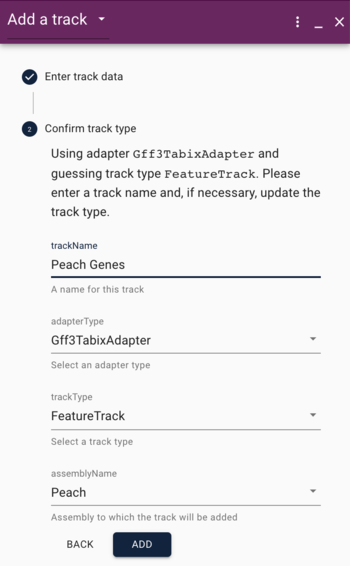
GFF (Generic Feature Format) is a very commonly used text format for describing features that exist on sequences. We'll head off to that page to talk about it a bit.
Features from a directory of files
Here, we'll use the Bio::DB::SeqFeature::Store adaptor in "memory" mode to read a directory of files. There are adaptors available for use with many other databases, such as Chado and Bio::DB::GFF.
Config file: pythium-1.conf
{
"description": "PAG 2017 P. ultima Example",
"db_adaptor": "Bio::DB::SeqFeature::Store",
"db_args" : {
"-adaptor" : "memory",
"-dir" : ".."
},
...