Difference between revisions of "Sybil Chado Comparative Genomics Data"
From GMOD
(New page: == Sybil Chado Comparative Genomics Data storage == This page will detail the TIGR/JCVI/IGS Sybil way of storing comparative genomics data. The following diagram describes how protein c...) |
(→Sybil Chado Comparative Genomics Data storage) |
||
| Line 1: | Line 1: | ||
| − | |||
== Sybil Chado Comparative Genomics Data storage == | == Sybil Chado Comparative Genomics Data storage == | ||
| Line 7: | Line 6: | ||
[[Image:ChadoSybilComparative.png]] | [[Image:ChadoSybilComparative.png]] | ||
| + | |||
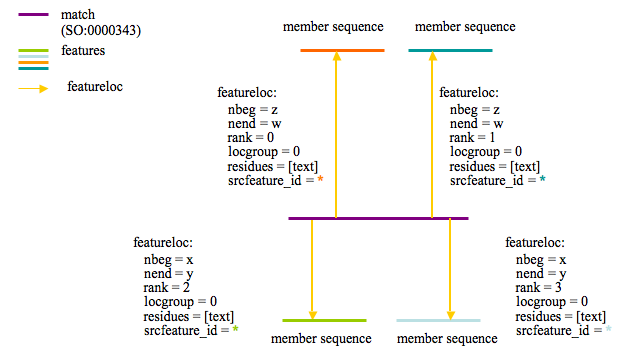
| + | The purple line represents the protein cluster and is stored in feature. | ||
| + | |||
| + | The proteins that are members of the cluster are shown with the labels 'member sequence' and are linked to the cluster feature via featurloc (where feature_id = protein feature and srcfeature_id = cluster feature). The featureloc rank column is incremented for each cluster member. | ||
Revision as of 17:09, 2 January 2008
Sybil Chado Comparative Genomics Data storage
This page will detail the TIGR/JCVI/IGS Sybil way of storing comparative genomics data.
The following diagram describes how protein clusters are stored for Sybil:
The purple line represents the protein cluster and is stored in feature.
The proteins that are members of the cluster are shown with the labels 'member sequence' and are linked to the cluster feature via featurloc (where feature_id = protein feature and srcfeature_id = cluster feature). The featureloc rank column is incremented for each cluster member.