Difference between revisions of "JBrowse"
m |
m (1 revision) |
||
| (88 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | <!-- to alter this page, please edit the raw data, which is stored at http://gmod.org/wiki/JBrowse/tool_data --> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | {{ :JBrowse/tool_data | template = Template:ToolDisplay }} | ||
| − | + | [[Category:GMOD Components]] | |
| − | + | [[Category:AJAX]] | |
| − | + | [[Category:JBrowse]] | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | [[ | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Revision as of 21:43, 13 February 2013
- Mature release
- Development: active
- Support: active
Included in
Contents
About JBrowse
JBrowse is a genome browser with a fully dynamic HTML5 user interface, being developed as the successor to GBrowse. It is very fast and scales well to large datasets. JBrowse does almost all of its work directly in the user's web browser, with minimal requirements for the server.
Features
- Fast, smooth scrolling and zooming. Explore your genome with unparalleled speed.
- Scales easily to multi-gigabase genomes.
- Supports GFF3, BED, FASTA, Wiggle, BigWig, BAM, and more.
- Very light server resource requirements. Serve huge datasets from a single low-cost cloud instance.
Visit the JBrowse website.
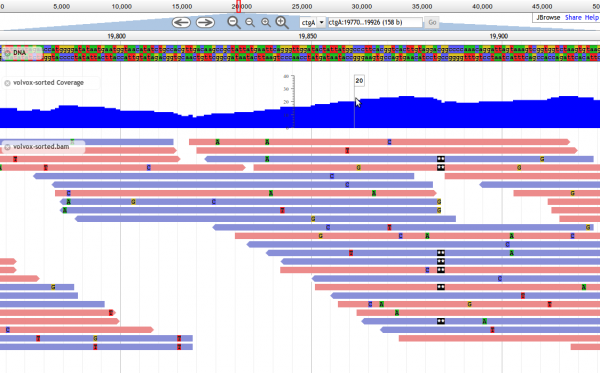
Screenshots
Downloads
- Download JBrowse: http://jbrowse.org/install/
- The source code for JBrowse can be downloaded from http://github.com/GMOD/jbrowse.
Using JBrowse
The JBrowse Quick-Start Tutorial provides a basic step-by-step recipe for quickly getting up and running with JBrowse.
System Requirements
JBrowse requires libpng, Zlib, and GD development libraries, plus make and a C compiler. On Ubuntu, you can install these prerequisites using the command:
sudo apt-get install libpng-dev libgd2-noxpm-dev build-essential
For tips on installing these baseline libraries, see JBrowse Troubleshooting.
Installation
The JBrowse Quick-Start Tutorial provides a basic step-by-step recipe for quickly getting up and running with JBrowse.
1. Download JBrowse onto your web server.
2. Unpack JBrowse into a directory that is served by your web browser. On many systems, this defaults to /var/www.
cd /var/www unzip JBrowse-*.zip
Make sure you have permissions to write to the contents of the jbrowse/ directory you have just created.
3. Run the automated-setup script, ./setup.sh, which will attempt to install all of JBrowse's (modest) prerequisites for you in the jbrowse/ directory itself. Note that setup.sh does not need to be run as root or with sudo. For help troubleshooting failures of setup.sh, see JBrowse Troubleshooting.
4. Visit JBrowse on your machine, substituting the http://(your_machine/path_to_jbrowse)/index.html?data=sample_data/json/volvox. If you can see the included Volvox example data, you are ready to configure JBrowse to show your own data!
Configuration
See the JBrowse Configuration Guide for information on:
- Formatting reference sequences (e.g. from FASTA files, or a Chado database)
- Feature Tracks (e.g. from BED or GFF files, a Chado database, or the UCSC genome browser)
- Image Tracks (e.g. from WIG files)
- Wiggle/BigWig Tracks
- Name Search and Autocompletion
- Removing tracks
- Compressing data stored on the server
- URL control
- Faceted track selection
- Anonymous usage statistics
Additional topics:
Upgrading JBrowse
To upgrade an existing JBrowse (1.3.0 or later) to the latest version, simply move its data directory (and jbrowse_conf.json if you are using it) into the directory of a newer JBrowse, and the new JBrowse will display that data.
To upgrade a 1.2.x JBrowse, copy its data directory into the new JBrowse directory, and point your browser at compat_121.html in the new JBrowse directory, instead of index.html.
If you are upgrading from a version of JBrowse older than 1.2.0, a fresh installation is required.
Publications, Tutorials, and Presentations
Tutorials
- JBrowse Tutorial covering installation and configuration
- part of the 2013 GMOD Summer School
- Getting Started with JBrowse Tutorial
- part of the JBrowse documentation
- Exploration of structural variation in the tomato clade using JBrowse
- Tutorial explaining how to browse structural variants from the 150+ tomato genome resequencing project (http://www.tomatogenome.net) using JBrowse
Presentations
- April 2013 - Bio-IT World, Robert Buels: PDF
- August 2012 - presentation given as part of the 2012 GMOD Summer School: PDF
- April 2012 - GMOD 2012 Community Meeting, Robert Buels: PDF
- January 2012 - Plant and Animal Genome (PAG) XX: PDF
- April 2010 - UCSC genome browser group ("genecats") meeting: PDF
- August 2009 - GMOD Community Meeting: Talk summary
Contacts and Mailing Lists
Please direct questions and inquiries regarding JBrowse to the mailing lists below.
Requests for help should be directed to gmod-ajax@lists.sourceforge.net.
| Mailing List Link | Description | Archive(s) | |
|---|---|---|---|
| JBrowse | gmod-ajax | JBrowse help and general questions. | Nabble (2010/05+), Sourceforge |
| jbrowse-dev | JBrowse development discussions. | Nabble (2011/08+) |
JBrowse Development
Development team
JBrowse is an open-source project, started and managed by the laboratory of Ian Holmes at the University of California, Berkeley.
As of January 2012, the lead developer of JBrowse is Robert Buels. Most of JBrowse was originally written by Mitch Skinner.
There is a mailing list for developers, and there is usually a teleconference on the 3rd Monday of the month at 2pm Pacific US time. We welcome participation from anyone and everyone. Please contact Robert Buels if you would like to listen in or participate.
Current status
The JBrowse source code repository is kept on GitHub. Please feel very free to fork the code on GitHub and make modifications and improvements, submitting pull requests. GitHub has a very nice tutorial on how to get started with this style of development.
More on JBrowse
See Category:JBrowse
Raw tool data at JBrowse/tool data