Difference between revisions of "GMOD Online Training 2014/WebApollo Tutorial"
(Created page with "==Using WebApollo== WebApollo is a web-based application, so the only requirement to use it is a web browser. It has been tested with Chrome, Firefox, and Safari. It has not...") |
|||
| Line 50: | Line 50: | ||
* Sequence searching | * Sequence searching | ||
| + | |||
| + | == Installing WebApollo == | ||
| + | |||
| + | ===Server operating system=== | ||
| + | Any Unix like system (e.g., Unix, Linux, Mac OS X) | ||
| + | |||
| + | ===Prerequisites=== | ||
| + | * Servlet container (must support servlet spec 3.0+) [officially supported: Tomcat 7] | ||
| + | * Relational Database Management System [officially supported: PostgreSQL] | ||
| + | * User database helper scripts | ||
| + | ** Perl modules | ||
| + | *** DBI | ||
| + | *** DBD module for your specific RDBMS [officially supported: DBD::Pg for PostgresSQL] | ||
| + | * Data generation pipeline (see [[JBrowse#Prerequisites|JBrowse prerequisites]] for more information on its prerequisites) | ||
| + | ** Perl modules | ||
| + | *** [[BioPerl|BioPerl 1.6]] | ||
| + | *** {{CPAN|JSON}} | ||
| + | *** {{CPAN|JSON::XS}} (optional, for speed) | ||
| + | *** {{CPAN|PerlIO::gzip}} | ||
| + | *** {{CPAN|Heap::Simple}} | ||
| + | *** {{CPAN|Heap::Simple::XS}} | ||
| + | *** {{CPAN|Devel::Size}} | ||
| + | *** {{CPAN|Hash::Merge}} | ||
| + | *** {{CPAN|Bio::GFF3::LowLevel::Parser}} | ||
| + | *** {{CPAN|Digest::Crc32}} | ||
| + | *** {{CPAN|Cache::Ref::FIFO}} | ||
| + | ** System packages | ||
| + | *** libpng12-0 | ||
| + | *** libpng12-dev | ||
| + | *** zlib1g (Debian/Ubuntu) | ||
| + | *** zlib1g-dev (Debian/Ubuntu) | ||
| + | *** zlib (RedHat/CentOS) | ||
| + | *** zlib-devel (RedHat/CentOS) | ||
| + | * Sequence search (optional) | ||
| + | ** Blat (along with a configured search database) | ||
| + | |||
| + | Note: All prerequisites have already been installed on the course's machine | ||
| + | |||
| + | === Installation === | ||
| + | |||
| + | The installation steps will be done in the command line. SSH into your AWS machine. | ||
| + | Uncompress the WebApollo-2014-04-03.tgz tarball. | ||
| + | |||
| + | $ <span class="enter">cd</span> | ||
| + | $ <span class="enter">tar -xvzf WebApollo-2014-04-03.tgz</span> | ||
| + | |||
| + | === User database === | ||
| + | WebApollo uses a database to determine who can access and edit annotations for a given sequence. | ||
| + | |||
| + | First we’ll need to create a database. You can call it whatever you want (remember the name as you’ll need to point the configuration to it). For the purposes of this guide, we’ll call it <tt>web_apollo_users</tt> You might want to create a separate account to manage the database. We’ll have the user <tt>web_apollo_db</tt> with password <tt>web_apollo_db</tt>. The user will not be a superuser nor will it be able to create new roles. But it will be able to create databases. | ||
| + | |||
| + | $ <span class="enter">createuser -P web_apollo_db | ||
| + | Enter password for new role: | ||
| + | Enter it again: | ||
| + | Shall the new role be a superuser? (y/n) n | ||
| + | Shall the new role be allowed to create databases? (y/n) y | ||
| + | Shall the new role be allowed to create more new roles? (y/n) n | ||
| + | </span> | ||
| + | |||
| + | Next we'll create the user database. | ||
| + | |||
| + | $ <span class="enter">createdb -U web_apollo_db web_apollo_users</span> | ||
| + | |||
| + | Now that the database is created, we need to load the schema to it. | ||
| + | |||
| + | $ <span class="enter">cd ~/WebApollo-2014-04-03/tools/user</span> | ||
| + | $ <span class="enter">psql -U web_apollo_db web_apollo_users < user_database_postgresql.sql</span> | ||
| + | |||
| + | Now the user database has been setup. | ||
| + | |||
| + | Let's populate the database. | ||
| + | |||
| + | First we’ll create an user with access to WebApollo. We’ll use the <tt>add_user.pl</tt> script in <tt>~/WebApollo-2014-04-03/tools/user</tt>. Let’s create an user named <tt>web_apollo_admin</tt> with the password <tt>web_apollo_admin</tt>. | ||
| + | |||
| + | $ <span class="enter">./add_user.pl -D web_apollo_users -U web_apollo_users_admin -P web_apollo_users_admin \ | ||
| + | -u web_apollo_admin -p web_apollo_admin</span> | ||
| + | |||
| + | Next we’ll add the annotation tracks ids for the genomic sequences for our organism. We’ll use the <tt>add_tracks.pl</tt> script in the same directory. We need to generate a file of genomic sequence ids for the script. For convenience, there’s a script called <tt>extract_seqids_from_fasta.pl</tt> in the same directory which will go through a FASTA file and extract all the ids from the deflines. Let’s first create the list of genomic sequence ids. We'll store it in <tt>~/scratch/seqids.txt</tt>. We’ll want to add the prefix “Annotations-” to each identifier. | ||
| + | |||
| + | $ <span class="enter">mkdir ~/scratch</span> | ||
| + | $ <span class="enter">./extract_seqids_from_fasta.pl -p Annotations- -i ~/maker_output/scf1117875582023.fa \ | ||
| + | -o ~/scratch/seqids.txt</span> | ||
| + | |||
| + | Now we’ll add those ids to the user database. | ||
| + | |||
| + | $ <span class="enter">./add_tracks.pl -D web_apollo_users -U web_apollo_db -P web_apollo_db \ | ||
| + | -t ~/scratch/seqids.txt</span> | ||
| + | |||
| + | Now that we have an user created and the annotation track ids loaded, we’ll need to give the user permissions to access the sequence. We’ll have the all permissions (read, write, publish, user manager). We’ll use the <tt>set_track_permissions.pl</tt> script in the same directory. We’ll need to provide the script a list of genomic sequence ids, like in the previous step. | ||
| + | |||
| + | $ <span class="enter">./set_track_permissions.pl -D web_apollo_users -U web_apollo_db \ | ||
| + | -P web_apollo_db -u web_apollo_admin -t ~/scratch/seqids.txt -a</span> | ||
| + | |||
| + | We’re all done setting up the user database. | ||
| + | |||
| + | Note that we’re only using a subset of the options for all the scripts mentioned above. You can get more detailed information on any given script (and other available options) using the “-h” or “--help” flag when running the script. | ||
Revision as of 18:51, 14 May 2014
Contents
Using WebApollo
WebApollo is a web-based application, so the only requirement to use it is a web browser. It has been tested with Chrome, Firefox, and Safari. It has not been tested with Internet Explorer.
A WebApollo demo with the Pythium data has been set up on the virtual machine. We'll use the demo to view WebApollo's functionality.
Point your browser to http://ec2-##-##-##-##.compute-1.amazonaws.com:8080/WebApolloDemo.
(e.g., http://ec2-184-73-92-243.compute-1.amazonaws.com:8080/WebApolloDemo).
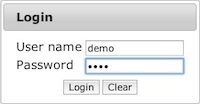
The user name and password are both demo.
We only have one contig to work with. Click on the scf1117875582023 link.
- Annotation track
- Add evidence tracks (maker, snap_masked, blastn, blastx, est2genome, protein2genome, BAM alignments)
- Moving around the contig
- Go to region scf1117875582023:629476..633770
- Selection
- Edge matching
- Create annotation
- Drag-n-drop
- Delete exon (notice the change in CDS)
- Add exon
- Non-canonical splice sites (GT / AG)
- Zoom to base
- DNA track
- Highlighting
- 6-frame translation
- DNA sequence insertion, deletion, substitution
- Zoom back out
- Change exon boundaries
- Merge (exons, transcripts)
- Split (exons, transcripts)
- Make intron
- Duplicate
- Set translation start
- Realtime client updating
- History
- Information editor
- Get sequence
- Sequence searching
Installing WebApollo
Server operating system
Any Unix like system (e.g., Unix, Linux, Mac OS X)
Prerequisites
- Servlet container (must support servlet spec 3.0+) [officially supported: Tomcat 7]
- Relational Database Management System [officially supported: PostgreSQL]
- User database helper scripts
- Perl modules
- DBI
- DBD module for your specific RDBMS [officially supported: DBD::Pg for PostgresSQL]
- Perl modules
- Data generation pipeline (see JBrowse prerequisites for more information on its prerequisites)
- Perl modules
- System packages
- libpng12-0
- libpng12-dev
- zlib1g (Debian/Ubuntu)
- zlib1g-dev (Debian/Ubuntu)
- zlib (RedHat/CentOS)
- zlib-devel (RedHat/CentOS)
- Sequence search (optional)
- Blat (along with a configured search database)
Note: All prerequisites have already been installed on the course's machine
Installation
The installation steps will be done in the command line. SSH into your AWS machine. Uncompress the WebApollo-2014-04-03.tgz tarball.
$ cd $ tar -xvzf WebApollo-2014-04-03.tgz
User database
WebApollo uses a database to determine who can access and edit annotations for a given sequence.
First we’ll need to create a database. You can call it whatever you want (remember the name as you’ll need to point the configuration to it). For the purposes of this guide, we’ll call it web_apollo_users You might want to create a separate account to manage the database. We’ll have the user web_apollo_db with password web_apollo_db. The user will not be a superuser nor will it be able to create new roles. But it will be able to create databases.
$ createuser -P web_apollo_db
Enter password for new role:
Enter it again:
Shall the new role be a superuser? (y/n) n
Shall the new role be allowed to create databases? (y/n) y
Shall the new role be allowed to create more new roles? (y/n) n
Next we'll create the user database.
$ createdb -U web_apollo_db web_apollo_users
Now that the database is created, we need to load the schema to it.
$ cd ~/WebApollo-2014-04-03/tools/user $ psql -U web_apollo_db web_apollo_users < user_database_postgresql.sql
Now the user database has been setup.
Let's populate the database.
First we’ll create an user with access to WebApollo. We’ll use the add_user.pl script in ~/WebApollo-2014-04-03/tools/user. Let’s create an user named web_apollo_admin with the password web_apollo_admin.
$ ./add_user.pl -D web_apollo_users -U web_apollo_users_admin -P web_apollo_users_admin \
-u web_apollo_admin -p web_apollo_admin
Next we’ll add the annotation tracks ids for the genomic sequences for our organism. We’ll use the add_tracks.pl script in the same directory. We need to generate a file of genomic sequence ids for the script. For convenience, there’s a script called extract_seqids_from_fasta.pl in the same directory which will go through a FASTA file and extract all the ids from the deflines. Let’s first create the list of genomic sequence ids. We'll store it in ~/scratch/seqids.txt. We’ll want to add the prefix “Annotations-” to each identifier.
$ mkdir ~/scratch $ ./extract_seqids_from_fasta.pl -p Annotations- -i ~/maker_output/scf1117875582023.fa \ -o ~/scratch/seqids.txt
Now we’ll add those ids to the user database.
$ ./add_tracks.pl -D web_apollo_users -U web_apollo_db -P web_apollo_db \
-t ~/scratch/seqids.txt
Now that we have an user created and the annotation track ids loaded, we’ll need to give the user permissions to access the sequence. We’ll have the all permissions (read, write, publish, user manager). We’ll use the set_track_permissions.pl script in the same directory. We’ll need to provide the script a list of genomic sequence ids, like in the previous step.
$ ./set_track_permissions.pl -D web_apollo_users -U web_apollo_db \
-P web_apollo_db -u web_apollo_admin -t ~/scratch/seqids.txt -a
We’re all done setting up the user database.
Note that we’re only using a subset of the options for all the scripts mentioned above. You can get more detailed information on any given script (and other available options) using the “-h” or “--help” flag when running the script.