GBrowse syn Database
GBrowse_syn is a GBrowse based synteny viewer. This page describes the database that GBrowse_syn uses, and how to get syntenic data into that database.
Contents
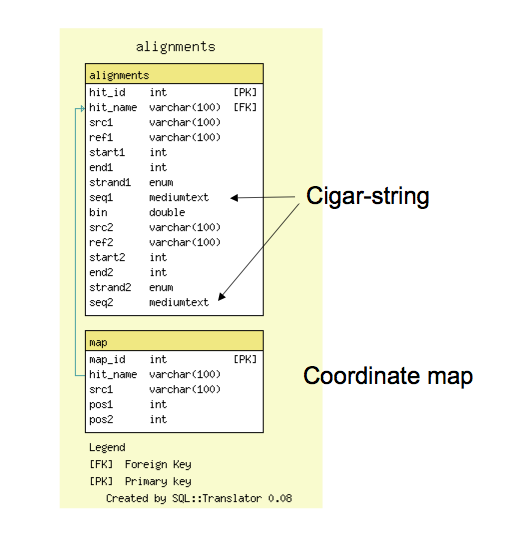
Schema
- The alignment database schema is very simple; it has a tables for all reciprocal 'hits', or alignment features and a table for (optional) 1:1 coordinate maps
- The alignments table contains coordinate information and also support cigar-line representations and the alignment to facilitate future reconstruction of the alignment within GBrowse_syn.
Sample data
- Sample data and configuration files can be downloaded from the GMOD FTP site.
- These data are for two rice species (courtesy of Bonnie Hurwitz)
Loading
Input formats for alignment, colinearity or synteny data
- The starting point for loading the alignment database is typically a multiple sequence alignment file
- The source of the alignment data is up to you but the supported aligment inputs formats for entry are currently CLUSTALW, FASTA, SELEX, MSF and STOCKHOLM. Which format is not particularly important, as long as is is recognized by the Bio::SeqIO library for parsing and allows sequence names long enough to accommodate the rules below.
NOTE: It is necessary to overload the sequence IDs with meta-data about the alignment, using naming convention "species-seqid(strand)/start-end" shown in the example below
Example Alignment Data
selex format
rice-3(+)/16598648-16598707 ggaggccggccgtctgccatgcgtgagccagacggggcgggccggagacaggccacgtgg wild_rice-3(+)/14467855-14467878 gggggccgg------------------------------------agacaggccacgtgg
MSF format
NoName MSF: 2 Type: N Thu Aug 6 10:56:43 2009 Check: 00 ..
Name: rice-3(+)/16598648-16598707 Len: 60 Check: 4764 Weight: 1.00
Name: wild_rice-3(+)/14467855-14467878 Len: 60 Check: 932 Weight: 1.00
//
1 50
rice-3(+)/16598648-16598707 ggaggccggc cgtctgccat gcgtgagcca gacggggcgg gccggagaca
wild_rice-3(+)/14467855-14467878 gggggccgg- ---------- ---------- ---------- -----agaca
51 100
rice-3(+)/16598648-16598707 ggccacgtgg
wild_rice-3(+)/14467855-14467878 ggccacgtgg
STOCKHOLM format
# STOCKHOLM 1.0 #=GF ID NoName #=GS rice-3(+)/16598648-16598707AC unknown #=GS wild_rice-3(+)/14467855-14467878AC unknown rice-3(+)/16598648-16598707 ggaggccggccgtctgccatgcgtgagccagacggggcgggccggagacaggccacgtgg wild_rice-3(+)/14467855-14467878 gggggccgg------------------------------------agacaggccacgtgg //
FASTA format
>rice-3(+)/16598648-16598707 ggaggccggccgtctgccatgcgtgagccagacggggcgggccggagacaggccacgtgg >wild_rice-3(+)/14467855-14467878 gggggccgg------------------------------------agacaggccacgtgg
CLUSTALW format
CLUSTAL W(1.81) multiple sequence alignment
rice-3(+)/16598648-16598707 ggaggccggccgtctgccatgcgtgagccagacggggcgggccggagacaggccacgtgg
wild_rice-3(+)/14467855-14467878 gggggccgg------------------------------------agacaggccacgtgg
** ******
Note on formats
These data are in clustalw and other format. The scripts used to process these data will recognize these commonly used formats recognized by BioPerl's AlignIO parser. This does not mean that clustalw or some other program are necessarily required to generate the alignment data.
- See this generalized hierarchical whole genome alignment workflow for general information on how whole genome alignment data ca be generated.
Note on the sequence ID syntax
The sequence ID is this clustal file is overloaded to contain information about the species, strand and coordinates. This information is essential:
rice-3(+)/16598648-16600199 speciesv-refseq(strand)/start-end
Database Loading Scripts
Split large clustal files into smaller ones
For performance reasons and to avoid overloading the bioperl-based perl scripts, split large clustal files that have many alignments into one alignment/file with the script split_clustal.pl. This is necessary for very large files that would otherwise overload the BioPerl alignment parser.
gunzip my_huge_clustal_file.aln.gz | perl split_clustal.pl /path/to/smaller_alignment_files
Parse the alignments
A suggested method for formatiing alignment data is to parse clustalw format the alignments using the script clustal2hit.pl
perl clustal2hit.pl /path/to/smaller_alignment_files/*.aln |gzip -c >processed_alignments.txt.gz
There is also a script for processing mercator alignments (mercatoraln_to_synhits.pl) and a generic version (aln2hit.pl) for other MSA formats supported by Bio::SeqIO;
- The output of this script is described below
alignment data loading format
- The output of the script is a tab-delimited ad hoc intermediate format that encodes the alignment coordinates plus 1:1 mapping of coordinates within the alignment (to facilitate accurate grid-lines in GBrowse_syn)
- However to get you alignment or syntency data, this the the format currently supported for loading the alignment database.
- Each record is a single line (wrapped here for display only)
#species1 seqid1 start1 end1 strand1 reserved species2 seqid2 start2 end2 strand2 reserved \ # pos1-1 pos1-2 ... posn-1 posn-2 | pos1-2 pos1-1 ... posn-2 posn-1 c_briggsae chrI 1583997 1590364 + . c_remanei Crem_Contig24 631879 634679 - . \ 1584000 634676 1584100 634584 (truncated...) | 631900 1590333 632000 1590233 (truncated ...)
Create a mysql database
mysql -uroot -ppassword -e 'create database my_database'
Load the database
load the database with the script load_alignment_database.pl
gunzip -c processed_alignments.txt.gz | perl load_alignment_database.pl
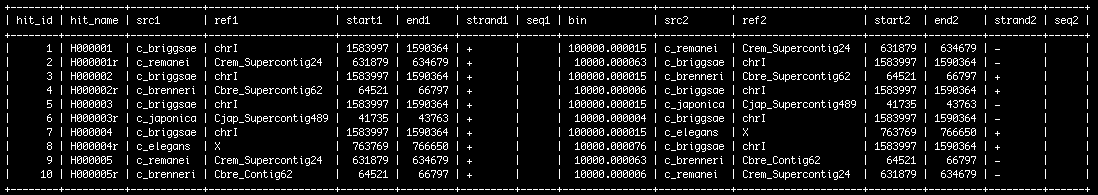
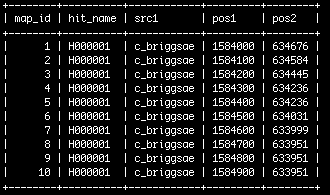
- Once loaded, the data look something like this: