Difference between revisions of "GBrowse syn"
From GMOD
| Line 1: | Line 1: | ||
<!--__NOEDITSECTION__--> | <!--__NOEDITSECTION__--> | ||
| − | |||
| − | |||
| − | |||
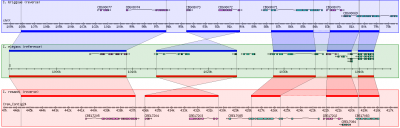
[[image:GBrowse_syn1_small.png|right|thumb|300px|GBrowse_syn running on WormBase]] | [[image:GBrowse_syn1_small.png|right|thumb|300px|GBrowse_syn running on WormBase]] | ||
| − | |||
{{GBrowse_syn_abstract}} | {{GBrowse_syn_abstract}} | ||
Revision as of 20:29, 16 July 2009
GBrowse_syn, or the Generic Synteny Browser, is a GBrowse-based synteny browser designed to display multiple genomes, with a central reference species compared to two or more additional species. It can be used to view multiple sequence alignment data, synteny or co-linearity data from other sources against genome annotations provided by GBrowse. GBrowse_syn is included with the standard GBrowse package (version 1.69 and later). Working examples can be seen at TAIR, WormBase, and SGN.
Contents
Documentation
Installation
- GBrowse_syn uses much of the same infrastructure as GBrowse, in terms of species databases, configuration files, perl libraries, etc.
- It comes as part of the GBrowse distribution, version 1.69 and later (GBrowse installation info...).
- It differs in that databases and configuration for individual species are linked together via a central configuration file and a joining database that contains reciprocal alignments between all species represented in the browser.
- We recommend using the most up-to-date version of the application either with the GBrowse netinstall script (use the -d option) or with a source code installation.
Configuration
- Configuration of GBrowse_syn is much the same as for GBrowse, with database and display options controlled by a configuration file
- GBrowse_syn uses a main configuration file for general options plus an individual configuration for each species represented in the mutliple sequence alignments
- Details of GBrowse_syn configuration can be found on this page.
The Alignment data
- GBrowse_syn uses a central 'joining' database that contains information about the multiple sequence alignments
- There is an additional database for each species represented in the alignments
- The species' databases are configured in the same way as a regular GBrowse installation
- Detailed information about the alignment database can be found on this page
User interface
- GBrowse_syn uses a central "reference species" panel, with inset panels above and below for two or more aligned species
- Details of the user interface can be found on this page
Presentations and Workshops
- Comparative Genomics with GBrowse_syn - Presentation by Sheldon McKay at the SMBE 2009 GMOD Workshop on using GBrowse_syn for comparative genomics.
- GBrowse_syn PAG 2009 Workshop
- November 2007 - Sheldon McKay's presentation on GBrowse_syn at the November 2007 GMOD Meeting.