Difference between revisions of "GBrowse img"
DanielRenfro (Talk | contribs) |
m (slight edit to clarify the 'source' argument) |
||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | '''gbrowse_img''' - CGI script to generate genome images via the [[Gbrowse|Generic Genome Browser]] | |
| − | + | ||
| − | '''gbrowse_img''' - CGI script to generate genome images via the Generic Genome Browser | + | |
__TOC__ | __TOC__ | ||
| + | |||
==Description== | ==Description== | ||
| − | + | This CGI script is an interface to the Generic Genome Browser for the purpose of retrieving dynamic images of a region of the genome. It can be used as the destination of an <img> tag like this: | |
<img src="http://heptamer.tamu.edu/cgi-bin/gb2/gbrowse_img/MG1655/?name=NC_000913:3923000..3951000"> | <img src="http://heptamer.tamu.edu/cgi-bin/gb2/gbrowse_img/MG1655/?name=NC_000913:3923000..3951000"> | ||
| − | The script can also be used to superimpose one or more external features onto the display, for example for the purpose of displaying BLAST hits, an STS or a knockout in the context of the genome. | + | The script can also be used to superimpose one or more external features onto the display, for example for the purpose of displaying BLAST hits, an STS or a knockout in the context of the genome. It is designed to use a URL-based API to draw or embed [[GBrowse]] images without loading the full genome browser interface. gbrowse_img can be used to embed Gbrowse images in other webpages or to create high-resolution images appropriate for publications. |
==Examples== | ==Examples== | ||
| − | + | Gbrowse 2.26 with the ''Escherichia coli'' K-12 MG1655 genome (from bp 3923000 to 3950999) has been used to generate the following figures. Also note that the URLs have been [http://en.wikipedia.org/wiki/Percent-encoding encoded]. Please be aware that your version of Gbrowse may operate slightly different based on its version. | |
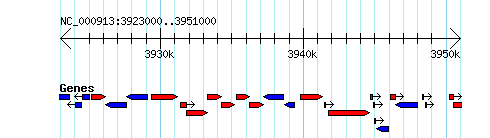
===Simple Example=== | ===Simple Example=== | ||
http://heptamer.tamu.edu/cgi-bin/gb2/gbrowse_img/MG1655/?name=NC_000913:3923000..3951000;l=Genes%1Elandmarks:overview%1EGenes:region%1Elandmarks:region;width=400;format=GD; | http://heptamer.tamu.edu/cgi-bin/gb2/gbrowse_img/MG1655/?name=NC_000913:3923000..3951000;l=Genes%1Elandmarks:overview%1EGenes:region%1Elandmarks:region;width=400;format=GD; | ||
| − | [[Image:Gbrowse img example2.png]] | + | |
| + | [[Image:Gbrowse img example2.png|border]] | ||
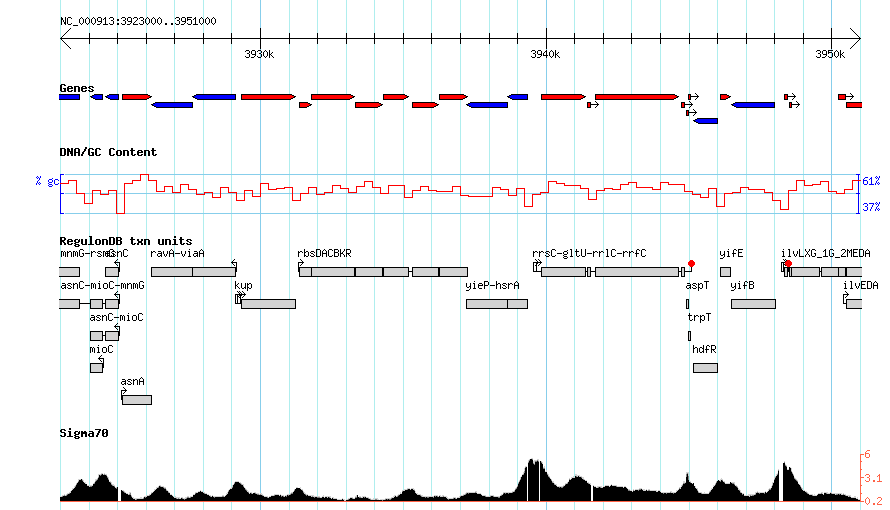
===More complex=== | ===More complex=== | ||
http://heptamer.tamu.edu/cgi-bin/gb2/gbrowse_img/MG1655/?name=NC_000913:3923000..3951000;l=Genes%1EDNA%1ERegulonDBtu%1ESigma70%1Elandmarks:overview%1EGenes:region%1Elandmarks:region;width=800;id=20736090abb824610d9d3bc89c8b4256;format=GD;keystyle=between;grid=1 | http://heptamer.tamu.edu/cgi-bin/gb2/gbrowse_img/MG1655/?name=NC_000913:3923000..3951000;l=Genes%1EDNA%1ERegulonDBtu%1ESigma70%1Elandmarks:overview%1EGenes:region%1Elandmarks:region;width=800;id=20736090abb824610d9d3bc89c8b4256;format=GD;keystyle=between;grid=1 | ||
| − | [[Image:Gbrowse img example1.png]] | + | |
| + | [[Image:Gbrowse img example1.png|border]] | ||
| + | |||
===Listing Sources=== | ===Listing Sources=== | ||
| Line 33: | Line 35: | ||
DH10B | DH10B | ||
MG1655 | MG1655 | ||
| − | . | + | ''etc.'' |
| + | ===Listing Types=== | ||
| + | To get a list of available tracks (the '''type''' parameter), set the '''list''' parameter to ''types'' like so: | ||
| + | http://heptamer.tamu.edu/cgi-bin/gb2/gbrowse_img/MG1655/?list=types | ||
| + | ## Feature types for source MG1655 | ||
| + | Genes Genes default | ||
| + | TranslationF 3-frame translation (forward) | ||
| + | TranslationR 3-frame translation (reverse) | ||
| + | DNA DNA/GC Content | ||
| + | Protein | ||
| + | rRNA_Operons | ||
| + | RegulonDBtu RegulonDB txn units default | ||
| + | Cryptic_Prophage cryptic prophage default | ||
| + | ''etc.'' | ||
==CGI arguments== | ==CGI arguments== | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
The script recognizes the following CGI arguments, which can be passed either as GET or POST argument=value pairs. Argument pairs must be separated by semicolons (preferred) or by ampersands. Many of the options have one-letter aliases that can be used to reduce URL lengths. | The script recognizes the following CGI arguments, which can be passed either as GET or POST argument=value pairs. Argument pairs must be separated by semicolons (preferred) or by ampersands. Many of the options have one-letter aliases that can be used to reduce URL lengths. | ||
| − | <table border="1"> | + | <table border="1" class="wikitable"> |
<tr><th>Argument</th><th>Alias</th><th>Description</th></tr> | <tr><th>Argument</th><th>Alias</th><th>Description</th></tr> | ||
<tr> <td>name</td> <td>q</td> <td>genomic landmark or range</td></tr> | <tr> <td>name</td> <td>q</td> <td>genomic landmark or range</td></tr> | ||
| + | <tr> <td>dbid</td> <td> </td> <td>database ID for disambiguating names</td></tr> | ||
<tr> <td>type</td> <td>t</td> <td>tracks to include in image</td></tr> | <tr> <td>type</td> <td>t</td> <td>tracks to include in image</td></tr> | ||
| Line 122: | Line 82: | ||
The arguments are explained in more detail here: | The arguments are explained in more detail here: | ||
| − | ; name | + | ; name / q |
: This argument specifies the region of the genome to be displayed. Several forms are recognized: | : This argument specifies the region of the genome to be displayed. Several forms are recognized: | ||
| − | + | :* ''name=Landmark'' | |
| − | + | :*: Display the landmark named "Landmark". Valid landmark names include chromosomes, contigs, clones, STSs, predicted genes, and any other landmark that the administrator has designated. Be careful when fetching large landmarks such as whole chromosomes! | |
| − | + | :* ''name=Landmark:start..end'' | |
| − | + | :*: Display the region between ''start'' and ''end'' relative to "Landmark". | |
| − | + | :* ''name=Class:Landmark'' | |
| − | + | :*: Display "Landmark", restricting to a particular class, such as "PCR_Product". The list of classes is under the control of the database administrator and is not yet available through this interface. | |
| + | :* ''name=Class:Landmark:start..end'' | ||
| + | :*: As above, but restricted to the designated range. | ||
| + | : If you use multiple <b>name</b> options, then this script will generate an overview image showing the position of each landmark. The alias "q" can be used to shorten the length of the URL. | ||
| − | + | ; dbid | |
| − | + | : If the data source contains multiple defined databases, this argument is required to uniquely identify landmarks that may appear in multiple databases under different names. If not present, the standard search algorithm is used. Use the symbolic database name indicated in the source configuration file. For example if the database stanza is "[scaffolds:database]" then pass "dbid=scaffolds". | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ; type /t | |
| − | ; type | + | |
: This argument lists the feature types to display. The value of this argument is a list of track names separated by spaces ("+" characters when L-escaped). For example: | : This argument lists the feature types to display. The value of this argument is a list of track names separated by spaces ("+" characters when L-escaped). For example: | ||
| − | + | : <pre>http://www.wormbase.org/db/seq/gbrowse_img/elegans?name=mec-3&type=tRNA+NG+WABA+CG+ESTB</pre> | |
| + | : Multiple ''type='' arguments will be combined to form a single space-delimited list. The alias "t" can be used to shorten the length of the URL. | ||
| + | : If the track name has a space in it, put quotes around the name: | ||
| + | : <pre>type="microbe tRNA"+NG+WABA+CG+ESTB</pre> | ||
| − | + | ; width / w | |
| − | + | : Width of the desired image, in pixels. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ; options / o | |
| − | + | : A space-delimited list ("+" characters when URL-escaped) of mnemonic/option pairs describing how features should be formatted. Options are integers from 0 to 3, where 0=auto, 1=compact, 2=expanded, 3=expanded and labeled. For example, to specify that the tRNA and NG tracks should always be expanded and labeled, but that the WABA track should be compact, use: | |
| − | + | : <pre>options=tRNA+3+NG+3+WABA+1</pre> | |
| − | + | : The alias "o" can be used to shorten the length of the URL. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ; add / a | |
| − | + | : Superimpose one or more additional features on top of the view. Features are specified as space ("+") delimited lists in the following format: | |
| − | + | : <pre>add=Landmark+Type+Name+start..end,start..end,start..end</pre> | |
| − | + | : "Landmark" is the landmark name, "Type" is a descriptive type that will be printed in the image caption, "Name" is a name for the feature to be printed above it, and start..end is a comma-delimited list of ranges for discontinuous feature. Names that contain white space must be quoted, for example "BLAST hit". Note that this all has to be URL-escaped, so an additional feature named "Your sequence", type "Blast Hit", that is located on chromosome III in a gapped range between 20000 and 22000, will be formatted as: | |
| − | + | : <pre>add=III+%22Blast%20Hit%22+%22Your%20Sequence%22+20000..21000,21550..22000</pre> | |
| − | + | : One or both of the type and name can be omitted. If omitted, type will default to "Your Features" and the name will default to "Feature XX" where XX is an integer. This allows for a very simple feature line: | |
| − | + | : <pre>add=III+20000..21000,21550..22000</pre> | |
| − | + | : Multiple ''add='' arguments are allowed. The alias "a" can be used to shorten the length of the URL. | |
| − | + | ||
| − | + | ||
| − | + | ; style | |
| − | + | : The style argument can be used to control the rendering of additional features added with "add". It is a flattened version of the style configuration sections described in [http://www.wormbase.org/db/seq/gbrowse?help=annotation this document]. For example, if you have added a "Blast Hit" annotation, then you can tell the renderer to use a red arrow for this glyph in this way: | |
| − | + | : <pre>style=%22Blast%20Hit%22+glyph=arrow+fgcolor=red</pre> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ; keystyle / k | |
| − | + | : Controls the positioning of the track key. One of "right", "left", "between" (default) or "bottom" | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ; overview | |
| − | + | : Ordinarily the image will show the detail panel if the query region corresponds to a single region, and the overview panel if multiple regions match (or if a region that is too large to show matches). Setting overview=1 will force the overview to be shown in all cases. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ; flip / f | |
| − | + | : Flip the image left to right. Arguments are 0=don't flip (default), and 1=flip. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ; embed | |
| − | + | : Generate image and a corresponding HTML imagemap in a form suitable for embedding into a frame. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ; format | |
| − | + | : Specify the format for the image file. Either "GD" (the default) or "GD::SVG" for scaleable vector graphics. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ; list | |
| − | + | : If this argument is present, it will cause the script to dump out various types of information in plain text form. Currently the two values for this argument are ''sources'', to dump out the list of data sources, and ''types'', to dump out the list of connfigured types. For ''list=sources'', the script will return a simple text list of the data source names. For ''list=types'', the script will return a three-column tab-delimited list giving the track names and feature types corresponding to the currently-selected data source. The format is as follows: | |
| − | + | : <pre>Mnemonic <tab> Full description of feature <tab> [default]</pre> | |
| − | + | : The third column contains the word "default" if the track will be shown by default when no ''type'' argument is provided. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ; source | |
| − | + | : This argument specifies the data source for the images. The list of sources can be found using ''list=sources''. See also the [[GBrowse_2.0_HOWTO#Configured_Data_Source_Sections configured data source sections]] in the GBrowse 2 guide. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ; h_feat | |
| − | <pre> | + | : The name of a feature to highlight in the format ''feature_name''@''color_name'' |
| − | < | + | : Example: |
| − | + | : <pre>h_feat=SKT5@blue</pre> | |
| + | : You may omit "@color", in which case the highlight will default to yellow. You can specify multiple h_feat arguments in order to highlight several features with distinct colors. | ||
| + | ; h_region | ||
| + | : The name of a region to highlight in a solid background color, in the format ''sequence_name'':''start''..''end''@''color_name'' | ||
| + | : Example: | ||
| + | : <pre>h_region=Chr3:200000..250000@wheat</pre> | ||
| + | : You may omit "@color", in which case the highlighted region will default to lightgrey. You can specify multiple h_region arguments in order to highlight several regions with distinct colors. | ||
| − | If you wish to associate the image with an imagemap so that clicking | + | ====Image-maps==== |
| − | on a feature takes the user to the destination configured in the | + | If you wish to associate the image with an imagemap so that clicking on a feature takes the user to the destination configured in the |
| − | gbrowse config file, you may do so by placing the URL in an | + | gbrowse config file, you may do so by placing the URL in an <iframe> section and using the ''embed=1'' flag: |
| − | <iframe> section and using the < | + | <pre><iframe src="http://localhost/cgi-bin/gbrowse_img/elegans?name=B0001;embed=1" width="100%" height="250"> |
| + | <img src="http://localhost/cgi-bin/gbrowse_img/elegans?name=B0001"/> | ||
| + | </iframe></pre> | ||
| + | Placing an <img> tag inside the <iframe> tag arranges for older browsers that don't know about iframes to display the static image instead. You may need to adjust the width and height attributes in order to avoid browsers placing scrollbars around the frame. | ||
| − | + | ==Generating from inside Gbrowse== | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
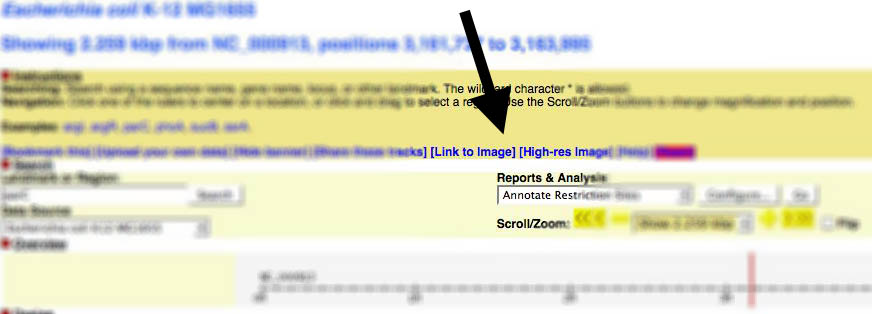
| − | + | ===Gbrowse v1.x=== | |
| + | You can find a link to generate images from within Gbrowse near the top of the page: | ||
| − | + | [[Image:Gbrowse img location v1.jpg|border]] | |
| − | + | ||
| − | image | + | |
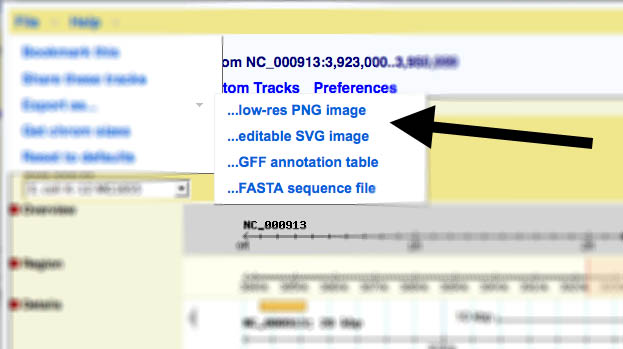
| − | + | ===Gbrowse v2.x=== | |
| + | In version 2, the link to generate images has been moved to the file menu: | ||
| + | |||
| + | [[Image:Gbrowse img location v2.jpg|border]] | ||
| + | |||
| + | |||
| + | ==Known Bugs== | ||
| + | The cookie that stores the configuration options for plugins does not transfer from gbrowse to gbrowse_img, so tracks generated by annotation plugins, such as the Restriction site annotator, will not display correctly when the image URL is generated on one machine and then viewed on another. Uploaded files will transfer correctly, however. | ||
| + | |||
| + | ==Author== | ||
| + | [mailto: lstein@cshl.org Lincoln Stein] | ||
| + | Copyright (c) 2002-2004 Cold Spring Harbor Laboratory | ||
| + | |||
| + | This library is free software; you can redistribute it and/or modify | ||
| + | it under the same terms as Perl itself. | ||
| + | |||
| + | ==Mediawiki Extension== | ||
| + | The [http://www.mediawiki.org/wiki/GBrowseImage GbrowseImage extension] for Mediawiki will display an image (rendered by gbrowse_img) in a wiki-page. | ||
| + | |||
| + | ==See Also== | ||
| + | * [[Gbrowse]] | ||
[[Category:Documentation]] | [[Category:Documentation]] | ||
[[Category:GBrowse]] | [[Category:GBrowse]] | ||
[[Category:GMOD Components]] | [[Category:GMOD Components]] | ||
Latest revision as of 22:02, 12 December 2012
gbrowse_img - CGI script to generate genome images via the Generic Genome Browser
Contents
Description
This CGI script is an interface to the Generic Genome Browser for the purpose of retrieving dynamic images of a region of the genome. It can be used as the destination of an <img> tag like this:
<img src="http://heptamer.tamu.edu/cgi-bin/gb2/gbrowse_img/MG1655/?name=NC_000913:3923000..3951000">
The script can also be used to superimpose one or more external features onto the display, for example for the purpose of displaying BLAST hits, an STS or a knockout in the context of the genome. It is designed to use a URL-based API to draw or embed GBrowse images without loading the full genome browser interface. gbrowse_img can be used to embed Gbrowse images in other webpages or to create high-resolution images appropriate for publications.
Examples
Gbrowse 2.26 with the Escherichia coli K-12 MG1655 genome (from bp 3923000 to 3950999) has been used to generate the following figures. Also note that the URLs have been encoded. Please be aware that your version of Gbrowse may operate slightly different based on its version.
Simple Example
More complex
Listing Sources
You can get a list of sources (genomes, chromosomes, etc.), by setting the list parameter to sources like so:
http://heptamer.tamu.edu/cgi-bin/gb2/gbrowse_img/MG1655/?list=sources
## Sources ATCC_8739 BL21 BL21_DE3 BL21_Gold_DE3 BW2952 DH10B MG1655 etc.
Listing Types
To get a list of available tracks (the type parameter), set the list parameter to types like so:
http://heptamer.tamu.edu/cgi-bin/gb2/gbrowse_img/MG1655/?list=types
## Feature types for source MG1655 Genes Genes default TranslationF 3-frame translation (forward) TranslationR 3-frame translation (reverse) DNA DNA/GC Content Protein rRNA_Operons RegulonDBtu RegulonDB txn units default Cryptic_Prophage cryptic prophage default etc.
CGI arguments
The script recognizes the following CGI arguments, which can be passed either as GET or POST argument=value pairs. Argument pairs must be separated by semicolons (preferred) or by ampersands. Many of the options have one-letter aliases that can be used to reduce URL lengths.
| Argument | Alias | Description |
|---|---|---|
| name | q | genomic landmark or range |
| dbid | database ID for disambiguating names | |
| type | t | tracks to include in image |
| width | w | desired width of image |
| options | o | list of track options (compact, labeled, etc) |
| abs | b | display position in absolute coordinates |
| add | a | added feature(s) to superimpose on the image |
| style | s | stylesheet for additional features |
| keystyle | k | where to place the image key |
| overview | force an overview-style display | |
| flip | f | flip image left to right |
| grid | turn grid on (1) or off (0) | |
| embed | generate full HTML for image and imagemap for use in an embedded frame | |
| format | format for the image (use "SVG" for scaleable vector graphics) | |
| list | get certain types of configuration information | |
| source | database name |
The arguments are explained in more detail here:
- name / q
- This argument specifies the region of the genome to be displayed. Several forms are recognized:
- name=Landmark
- Display the landmark named "Landmark". Valid landmark names include chromosomes, contigs, clones, STSs, predicted genes, and any other landmark that the administrator has designated. Be careful when fetching large landmarks such as whole chromosomes!
- name=Landmark:start..end
- Display the region between start and end relative to "Landmark".
- name=Class:Landmark
- Display "Landmark", restricting to a particular class, such as "PCR_Product". The list of classes is under the control of the database administrator and is not yet available through this interface.
- name=Class:Landmark:start..end
- As above, but restricted to the designated range.
- name=Landmark
- If you use multiple name options, then this script will generate an overview image showing the position of each landmark. The alias "q" can be used to shorten the length of the URL.
- dbid
- If the data source contains multiple defined databases, this argument is required to uniquely identify landmarks that may appear in multiple databases under different names. If not present, the standard search algorithm is used. Use the symbolic database name indicated in the source configuration file. For example if the database stanza is "[scaffolds:database]" then pass "dbid=scaffolds".
- type /t
- This argument lists the feature types to display. The value of this argument is a list of track names separated by spaces ("+" characters when L-escaped). For example:
-
http://www.wormbase.org/db/seq/gbrowse_img/elegans?name=mec-3&type=tRNA+NG+WABA+CG+ESTB
- Multiple type= arguments will be combined to form a single space-delimited list. The alias "t" can be used to shorten the length of the URL.
- If the track name has a space in it, put quotes around the name:
-
type="microbe tRNA"+NG+WABA+CG+ESTB
- width / w
- Width of the desired image, in pixels.
- options / o
- A space-delimited list ("+" characters when URL-escaped) of mnemonic/option pairs describing how features should be formatted. Options are integers from 0 to 3, where 0=auto, 1=compact, 2=expanded, 3=expanded and labeled. For example, to specify that the tRNA and NG tracks should always be expanded and labeled, but that the WABA track should be compact, use:
-
options=tRNA+3+NG+3+WABA+1
- The alias "o" can be used to shorten the length of the URL.
- add / a
- Superimpose one or more additional features on top of the view. Features are specified as space ("+") delimited lists in the following format:
-
add=Landmark+Type+Name+start..end,start..end,start..end
- "Landmark" is the landmark name, "Type" is a descriptive type that will be printed in the image caption, "Name" is a name for the feature to be printed above it, and start..end is a comma-delimited list of ranges for discontinuous feature. Names that contain white space must be quoted, for example "BLAST hit". Note that this all has to be URL-escaped, so an additional feature named "Your sequence", type "Blast Hit", that is located on chromosome III in a gapped range between 20000 and 22000, will be formatted as:
-
add=III+%22Blast%20Hit%22+%22Your%20Sequence%22+20000..21000,21550..22000
- One or both of the type and name can be omitted. If omitted, type will default to "Your Features" and the name will default to "Feature XX" where XX is an integer. This allows for a very simple feature line:
-
add=III+20000..21000,21550..22000
- Multiple add= arguments are allowed. The alias "a" can be used to shorten the length of the URL.
- style
- The style argument can be used to control the rendering of additional features added with "add". It is a flattened version of the style configuration sections described in this document. For example, if you have added a "Blast Hit" annotation, then you can tell the renderer to use a red arrow for this glyph in this way:
-
style=%22Blast%20Hit%22+glyph=arrow+fgcolor=red
- keystyle / k
- Controls the positioning of the track key. One of "right", "left", "between" (default) or "bottom"
- overview
- Ordinarily the image will show the detail panel if the query region corresponds to a single region, and the overview panel if multiple regions match (or if a region that is too large to show matches). Setting overview=1 will force the overview to be shown in all cases.
- flip / f
- Flip the image left to right. Arguments are 0=don't flip (default), and 1=flip.
- embed
- Generate image and a corresponding HTML imagemap in a form suitable for embedding into a frame.
- format
- Specify the format for the image file. Either "GD" (the default) or "GD::SVG" for scaleable vector graphics.
- list
- If this argument is present, it will cause the script to dump out various types of information in plain text form. Currently the two values for this argument are sources, to dump out the list of data sources, and types, to dump out the list of connfigured types. For list=sources, the script will return a simple text list of the data source names. For list=types, the script will return a three-column tab-delimited list giving the track names and feature types corresponding to the currently-selected data source. The format is as follows:
-
Mnemonic <tab> Full description of feature <tab> [default]
- The third column contains the word "default" if the track will be shown by default when no type argument is provided.
- source
- This argument specifies the data source for the images. The list of sources can be found using list=sources. See also the GBrowse_2.0_HOWTO#Configured_Data_Source_Sections configured data source sections in the GBrowse 2 guide.
- h_feat
- The name of a feature to highlight in the format feature_name@color_name
- Example:
-
h_feat=SKT5@blue
- You may omit "@color", in which case the highlight will default to yellow. You can specify multiple h_feat arguments in order to highlight several features with distinct colors.
- h_region
- The name of a region to highlight in a solid background color, in the format sequence_name:start..end@color_name
- Example:
-
h_region=Chr3:200000..250000@wheat
- You may omit "@color", in which case the highlighted region will default to lightgrey. You can specify multiple h_region arguments in order to highlight several regions with distinct colors.
Image-maps
If you wish to associate the image with an imagemap so that clicking on a feature takes the user to the destination configured in the gbrowse config file, you may do so by placing the URL in an <iframe> section and using the embed=1 flag:
<iframe src="http://localhost/cgi-bin/gbrowse_img/elegans?name=B0001;embed=1" width="100%" height="250"> <img src="http://localhost/cgi-bin/gbrowse_img/elegans?name=B0001"/> </iframe>
Placing an <img> tag inside the <iframe> tag arranges for older browsers that don't know about iframes to display the static image instead. You may need to adjust the width and height attributes in order to avoid browsers placing scrollbars around the frame.
Generating from inside Gbrowse
Gbrowse v1.x
You can find a link to generate images from within Gbrowse near the top of the page:
Gbrowse v2.x
In version 2, the link to generate images has been moved to the file menu:
Known Bugs
The cookie that stores the configuration options for plugins does not transfer from gbrowse to gbrowse_img, so tracks generated by annotation plugins, such as the Restriction site annotator, will not display correctly when the image URL is generated on one machine and then viewed on another. Uploaded files will transfer correctly, however.
Author
[mailto: lstein@cshl.org Lincoln Stein] Copyright (c) 2002-2004 Cold Spring Harbor Laboratory
This library is free software; you can redistribute it and/or modify it under the same terms as Perl itself.
Mediawiki Extension
The GbrowseImage extension for Mediawiki will display an image (rendered by gbrowse_img) in a wiki-page.