Difference between revisions of "Creating and Managing Subtracks with GBrowse2"
(→A Basic Example) |
(→A Basic Example) |
||
| Line 13: | Line 13: | ||
key = Wormbase Genes | key = Wormbase Genes | ||
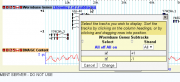
| − | [[image:subtracks_genes_before.png| | + | [[image:subtracks_genes_before.png|thumb|right|Subtrack selector]] |
The behavior of this track is to show both forward and reverse strand genes packed together for maximum efficiency, as shown in the figure on the right. Let's say we would prefer for forward and reverse stranded genes to be sorted into separate subtracks so that they do not intermingle. This can be done by adding '''subtrack select''' and '''subtrack table''' options to the configuration: | The behavior of this track is to show both forward and reverse strand genes packed together for maximum efficiency, as shown in the figure on the right. Let's say we would prefer for forward and reverse stranded genes to be sorted into separate subtracks so that they do not intermingle. This can be done by adding '''subtrack select''' and '''subtrack table''' options to the configuration: | ||
Revision as of 17:28, 14 June 2010
As of GBrowse version 2.09, you can create an unlimited number of subtracks within a single major track in order to group a series of datasets that are logically linked, such as a timecourse. You can choose which subtracks to show by default and the order in which they will appear. When the user clicks a designated area in the titlebar, a dialog box appears that allows the user to select which subtracks to make visible. The user can also drag subtrack labels up and down to adjust the order in which they are displayed.
A Basic Example
Here is a simple example to show how the system works. We start out with a gene track that has no subtracks:
[Genes] feature = gene glyph = gene database = sqlite-genes category = Genes:Coding label = 1 key = Wormbase Genes
The behavior of this track is to show both forward and reverse strand genes packed together for maximum efficiency, as shown in the figure on the right. Let's say we would prefer for forward and reverse stranded genes to be sorted into separate subtracks so that they do not intermingle. This can be done by adding subtrack select and subtrack table options to the configuration:
[Genes]
feature = gene
glyph = gene
database = sqlite-genes
category = Genes:Coding
label = 1
subtrack select = Strand strand
subtrack table = +1 ;
-1
key = Wormbase Genes
The "subtrack select" option defines a partitioning scheme for the data in the track. It consists of one or more lines defining the dimensions on which to partition the data. In this case, we are partitioning on only one dimension, the strandedness of the feature. The definition of this dimension is <Dimension Label> <method> (whitespace delimited, as usual), where the dimension label is a human-readable column label on the selection dialog and the method is any of the methods recognized by Bio::SeqFeatureI objects ("display_name", "primary_tag", "source_tag", "score", "has_tag", "tag_value", "start", "length", etc). In the example above, we are going to partition on the strand() method, and to label this dimension "Strand" on the popup menu presented to the user.
The "subtrack table" option defines the values on which to partition the data. In this case, we are going to partition the data into two subtracks: one for positive strand features (strand() returns +1) and one for negative strand features (strand() returns -1).
Reloading the browser now gives the track shown on the upper right. When the user clicks on the titlebar where it says "Showing 2 of 2 tracks", he can control the display of the subtracks using the dialog on the lower right.
Wiggle Track Example
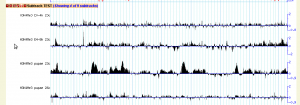
The image on the right shows an excerpt from the histone modification ChIP-seq track from the modENCODE project and the dialog box used to select among the subtracks. This track was created with the following configuration:
[ChIP-seq]
subtrack select = "Antibody" tag_value antibody ;
"Confirmed" has_tag confirmed ;
"Stage" tag_value stage ;
"Temperature" tag_value temp ;
subtrack table = H3K4Me3 1 E0-4h 23 =1 * ;
H3K4Me3 0 E4-8h 23 =2 ;
H3K4Me3 0 pupae 23 =3 ;
H3K4Me3 0 pupae 26 =4 ;
H3K9Me2 0 E0-4h 23 =5 * ;
H3K9Me2 1 E4-8h 23 =6 ;
H3K9Me2 0 pupae 23 =7 ;
H3K9Me2 1 pupae 26 =8 ;
H3K27Me3 1 E0-4h 23 =9 * ;
brief comment = This track shows modENCODE ChIP-seq characterization of
histone marks across various stages and growth conditions.
subtrack labels = E0-4h "Early embryo" ;
E4-8h "Late embryo" ;
pupae "Pupating larvae" ;
The following options are used here:
- subtrack select
- This required option defines the dimensions along which the subtracks are defined. The combination of dimensions are used to create the column headings for each subtrack. Each dimension consists of three white-space separated values, terminated by a semicolon. The first value is the column label, e.g. "Antibody". The second column is a method call name to be applied to the feature (discussed at more length later). The optional third argument is an argument to be passed to the method call.
The method name can be any of the methods accepted by Bio::SeqFeatureI feature objects. Typical methods used for the second argument are "display_name" (the name of the feature), "type", "method", "source", "score", "has_tag", and "tag_value". "has_tag" and "tag_value" both require the third argument to determine which tag to interrogate. For example, in the example above, the "Antibody" column is populated by interrogating each feature by calling $feature->tag_value('antibody').
- subtrack table
- This required option defines the subtracks; one line for each row of the subtracks selection table. Each line is terminated by a semicolon (the very last line doesn't actually need to be terminated). Within each line are a series of values to be matched against the dimensions specified by "subtrack select". In the above example, the first subtrack is defined by features whose antibody tag equals "H3K4Me3", has the tag "confirmed", has a stage tag matching "E0-4h" and a "temp" tag matching "23". The second subtrack is defined by features whose antibody tag equals "H3K4Me3", does not have the tag "confirmed", whose stage matches "E4-8h", and so on. There must be exactly as many match values in each row of "subtrack table" as there are dimensions defined in "subtrack select." Matching is done using a case-sensitive exact match unless you preface the value with a "~" sign, in which case a case-insensitive regular expression match is used.
Additional arguments can follow the match values. An argument that begins with the "=" sign assigns an ID to the subtrack. This ID can be used to turn the subtrack on and off via the URL-based API. For example, calling GBrowse's URL with the arguments "?l=ChIP-seq/1,2,5" will turn on this track's subtracks 1, 2 and 5. An asterisk (*) specifies which tracks are turned on by default. The ID and the asterisk, if any, can occur anywhere in the subtrack definition line.
- brief comment
- This optional option is a bit of explanatory text to insert at the top of the subtrack selection dialog.
- subtrack labels
If you would like to select subtracks based on its type, a tag, or some other method, but you want the user to see a different label for the value when it is displayed in the subtrack selection dialog, you can specify the label here. In the example above, we are selecting certain subtracks based on their developmental stage, but instead of making the internal names for the stages visible (E0-4h, E4-8h, etc), we ask GBrowse to display more friendly names ("Early embryo", "Late embryo", etc.)
Finally, here is a simple one-dimensional example that defines several subtracks based on their display names:
[ExpressionArrays]
subtrack select = "Array Name" display_name
subtrack table = Affy200K ;
Affy600K ;
AffyCpG28 ;
and here is another example that is functionally identical to the example given for the deprecated "select" option. Note the use of the ~ to indicate that the matches are to use case-insensitive regular expressions.
[GrowthCurve]
subtrack select = "Sample Time" source
subtrack table = '~day 1' ;
'~day 2' ;
'~day 3'